

新疆农业科学 ›› 2024, Vol. 61 ›› Issue (3): 521-536.DOI: 10.6048/j.issn.1001-4330.2024.03.001
• 作物遗传育种·种质资源·分子遗传学·生理生化 • 上一篇 下一篇
王凯迪1( ), 高晨旭2, 裴文锋1,2, 杨书贤2, 张文庆2, 宋吉坤1,2, 马建江2, 王莉2, 于霁雯1,2(
), 高晨旭2, 裴文锋1,2, 杨书贤2, 张文庆2, 宋吉坤1,2, 马建江2, 王莉2, 于霁雯1,2( ), 陈全家1(
), 陈全家1( )
)
收稿日期:2023-07-11
出版日期:2024-03-20
发布日期:2024-04-19
通信作者:
于霁雯(1978-),女,河南安阳人,研究员,博士,硕士生/博士生导师,研究方向为棉花分子育种,(E-mail)yujw666@hotmail.com;作者简介:王凯迪(1998-),男,陕西延安人,硕士研究生,研究方向为棉花分子育种,(E-mail)424416709@qq.com
基金资助:
WANG Kaidi1( ), GAO Chenxu2, PEI Wenfeng1,2, YANG Shuxian2, ZHANG Wenqing2, SONG Jikun1,2, MA Jianjiang2, WANG Li2, YU Jiwen1,2(
), GAO Chenxu2, PEI Wenfeng1,2, YANG Shuxian2, ZHANG Wenqing2, SONG Jikun1,2, MA Jianjiang2, WANG Li2, YU Jiwen1,2( ), CHEN Quanjia1(
), CHEN Quanjia1( )
)
Received:2023-07-11
Published:2024-03-20
Online:2024-04-19
Supported by:摘要:
【目的】 鉴定陆地棉TRM基因家族序列及理化性质,分析与棉纤维品质相关的优异单倍型基因差异。【方法】 利用生物信息学分析陆地棉TRM基因家族进化关系、理化性质和聚类表达;利用基因单倍型效应分析筛选调控纤维品质性状(纤维长度、比强度、马克隆值)的候选基因。【结果】 陆地棉TRM基因家族编码的氨基酸为376~1 093,等电点为4.64~9.56。亚细胞定位预测发现多达60个陆地棉TRM基因家族定位于细胞核中。TRM基因家族含有较多的光响应、激素响应、胁迫响应和生长发育相关的元件。60个TRM基因家族在纤维发育时期优势表达,调控棉花纤维发育。每个基因的单倍型个数为1~8,并筛选到纤维长度、强度以及马克隆值相关的优异单倍型TRM基因分别有14、18和15个,其中有11个基因同时具有长度、强度、马克隆值3种改良纤维品质的优异单倍型。GH_D09G0775的Hap_4单倍型和GH_D03G1434的Hap_3单倍型在增加纤维长度、强度的同时降低了马克隆值。【结论】 在陆地棉(Gossypium hirsutum L.)中鉴定出75个TRM基因家族成员分布在24条染色体上,系统进化将其分为Cluster Ⅰ~Ⅲ 3个亚族。
中图分类号:
王凯迪, 高晨旭, 裴文锋, 杨书贤, 张文庆, 宋吉坤, 马建江, 王莉, 于霁雯, 陈全家. 陆地棉TRM基因家族的鉴定及纤维品质相关优异单倍型分析[J]. 新疆农业科学, 2024, 61(3): 521-536.
WANG Kaidi, GAO Chenxu, PEI Wenfeng, YANG Shuxian, ZHANG Wenqing, SONG Jikun, MA Jianjiang, WANG Li, YU Jiwen, CHEN Quanjia. Identification of TRM gene family and fiber quality related excellent haplotype analysis in Gossypium hirsutum L.[J]. Xinjiang Agricultural Sciences, 2024, 61(3): 521-536.
| 基因ID Gene ID | 转录本长度 Transcript Length(bp) | 染色体 Chromo- some | 基因组位置 Genomic Location | 氨基酸数量 Number of Amino Acid | 分子量 Molecular Weight | 等电点 pI | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| GH_A01G1410.1 | 1 314 | A01 | 54808764-54811032 | 437 | 49.92686 | 4.88 | nucl |
| GH_A01G2253.1 | 1 518 | A01 | 115594835-115597597 | 505 | 57.79641 | 4.86 | nucl |
| GH_A01G2363.1 | 2 700 | A01 | 116832063-116835585 | 899 | 100.41775 | 6.11 | cyto |
| GH_A02G0234.1 | 2 757 | A02 | 2404926-2408330 | 918 | 103.50754 | 5.22 | nucl |
| GH_A02G2044.1 | 2 625 | A02 | 108074808-108078882 | 874 | 99.90307 | 6.04 | nucl |
| GH_A03G0533.1 | 1 563 | A03 | 8153312-8155305 | 520 | 60.28175 | 5.68 | nucl |
| GH_A03G1103.1 | 2 793 | A03 | 50408499-50412418 | 930 | 105.21887 | 5.07 | nucl |
| GH_A04G0979.1 | 1 434 | A04 | 69462180-69469536 | 477 | 52.62433 | 4.64 | nucl |
| GH_A05G0964.1 | 2 346 | A05 | 8607869-8610697 | 781 | 88.96019 | 6.01 | nucl |
| GH_A05G3931.1 | 3 123 | A05 | 102844278-102849879 | 1040 | 116.20696 | 9.31 | nucl |
| GH_A07G0059.1 | 1 533 | A07 | 628988-631260 | 510 | 58.37572 | 8.9 | nucl |
| GH_A07G0755.1 | 3 282 | A07 | 9022671-9027515 | 1093 | 120.99027 | 8.98 | nucl |
| GH_A07G2291.1 | 2 961 | D07 | 91905861-91909976 | 986 | 110.2077 | 6.3 | nucl |
| GH_A07G2350.1 | 2 331 | A07 | 92889383-92892178 | 776 | 87.42472 | 9.27 | nucl |
| GH_A08G0692.1 | 2 229 | A08 | 10127217-10130953 | 742 | 83.86942 | 9.19 | nucl |
| GH_A08G1551.1 | 1 131 | A08 | 102402502-102404142 | 376 | 42.99415 | 5.11 | nucl |
| GH_A08G2046.1 | 1 428 | A08 | 115647936-115650980 | 475 | 54.38497 | 5.52 | nucl |
| GH_A08G2091.1 | 1 770 | A08 | 116652133-116659356 | 589 | 67.4778 | 9.15 | chlo |
| GH_A08G2193.1 | 3 021 | A08 | 117970079-117975125 | 1006 | 112.14071 | 6.14 | nucl |
| GH_A09G0827.1 | 2 682 | A09 | 56330011-56333092 | 893 | 99.83975 | 9.11 | nucl |
| GH_A09G1541.1 | 1 524 | A09 | 71774616-71777153 | 507 | 57.473 | 5.23 | nucl |
| GH_A09G1756.1 | 2 982 | A09 | 74454345-74458907 | 993 | 110.9183 | 6.16 | nucl |
| GH_A09G1944.1 | 2 085 | A09 | 76570427-76572832 | 694 | 78.50671 | 9.56 | vacu |
| GH_A09G2222.1 | 2 697 | A09 | 79100072-79103507 | 898 | 101.38568 | 5.71 | chlo |
| GH_A09G2652.1 | 3 216 | A09 | 82737688-82741940 | 1071 | 119.47197 | 9.14 | nucl |
| GH_A10G1840.1 | 2 709 | A10 | 96909927-96913212 | 902 | 100.12998 | 9.1 | nucl |
| GH_A10G2325.1 | 2 406 | A10 | 109893241-109895998 | 801 | 91.39894 | 5.62 | nucl |
| GH_A11G0638.1 | 2 541 | A11 | 5673205-5676085 | 846 | 93.87066 | 9.49 | nucl |
| GH_A11G1309.1 | 2 355 | A11 | 13852756-13856249 | 784 | 88.58708 | 9.3 | nucl |
| GH_A11G1709.1 | 2 916 | A11 | 21197533-21201779 | 971 | 107.75294 | 8.55 | chlo |
| GH_A11G3051.1 | 2 604 | A11 | 111002265-111006157 | 867 | 96.23437 | 8.47 | nucl |
| GH_A11G3723.1 | 2 442 | A11 | 121125176-121128740 | 813 | 92.20752 | 5.62 | chlo |
| GH_A12G2452.1 | 1 251 | A12 | 102934223-102937012 | 416 | 47.26911 | 7.6 | nucl |
| GH_A12G2487.1 | 2 748 | A12 | 103276025-103279440 | 915 | 103.29828 | 8.32 | mito |
| GH_A12G2819.1 | 2 136 | A12 | 106188338-106190738 | 711 | 79.65164 | 9.26 | nucl |
| GH_A13G1327.1 | 2 727 | A13 | 78846901-78851063 | 908 | 101.54571 | 6.84 | nucl |
| GH_D01G1490.1 | 1 308 | D01 | 31939006-31941324 | 435 | 49.87765 | 4.87 | nucl |
| GH_D01G2334.1 | 1 515 | D01 | 62367964-62370708 | 504 | 57.81324 | 4.8 | nucl |
| GH_D01G2442.1 | 2 700 | D01 | 63514690-63518172 | 899 | 100.38982 | 5.99 | cyto |
| GH_D02G0253.1 | 2 760 | D02 | 2744183-2747593 | 919 | 103.89322 | 5.33 | mito |
| GH_D02G0972.1 | 1 272 | D02 | 20372804-20374278 | 423 | 48.19872 | 9.06 | nucl |
| GH_D02G1228.1 | 2 793 | D02 | 35813214-35817125 | 930 | 105.24384 | 4.93 | nucl |
| GH_D03G0017.1 | 2 625 | D03 | 138690-142736 | 874 | 99.91123 | 6.13 | nucl |
| GH_D03G1434.1 | 1 563 | D03 | 46317807-46319773 | 520 | 59.66096 | 5.74 | nucl |
| GH_D04G0442.1 | 3 126 | D04 | 6606757-6612330 | 1041 | 116.1618 | 9.36 | nucl |
| GH_D04G1311.1 | 2 853 | D04 | 43160828-43164823 | 950 | 105.2462 | 5.81 | nucl |
| GH_D05G0951.1 | 2 331 | D05 | 7672457-7675301 | 776 | 88.00605 | 6.03 | nucl |
| GH_D05G3052.1 | 2 688 | D05 | 31538189-31546485 | 895 | 102.41937 | 6.85 | nucl |
| GH_D07G0068.1 | 2 217 | D07 | 603139-606111 | 738 | 84.33982 | 9.02 | nucl |
| GH_D07G0751.1 | 3 279 | D07 | 8003392-8008211 | 1092 | 120.65422 | 9.27 | nucl |
| GH_D07G1883.1 | 2 877 | D07 | 38696495-38700181 | 958 | 109.3117 | 7.24 | nucl |
| GH_D07G2236.1 | 2 904 | D07 | 53803702-53807843 | 967 | 107.98503 | 5.97 | nucl |
| GH_D07G2293.1 | 1 923 | D07 | 54683888-54686682 | 640 | 72.50937 | 9.54 | nucl |
| GH_D08G0689.1 | 2 229 | D08 | 8659310-8663064 | 742 | 83.75555 | 9.36 | nucl |
| GH_D08G1569.1 | 1 131 | D08 | 51012741-51014358 | 376 | 42.8249 | 5.1 | nucl |
| GH_D08G2058.1 | 1 428 | D08 | 60460228-60462948 | 475 | 54.34586 | 5.45 | nucl |
| GH_D08G2107.1 | 1 725 | D08 | 61169567-61171897 | 574 | 65.66284 | 9.43 | chlo |
| GH_D08G2206.1 | 3 015 | D08 | 62410993-62416003 | 1004 | 112.23577 | 6.18 | nucl |
| GH_D09G0775.1 | 2 682 | D09 | 30764289-30767370 | 893 | 99.69663 | 9.03 | nucl |
| GH_D09G1524.1 | 1 548 | D09 | 42192153-42194698 | 515 | 58.35159 | 5 | nucl |
| GH_D09G1704.1 | 2 970 | D09 | 44087946-44092487 | 989 | 110.36687 | 6.26 | nucl |
| GH_D09G1898.1 | 2 070 | D09 | 46163346-46165731 | 689 | 77.82586 | 9.46 | vacu |
| GH_D09G2155.1 | 2 697 | D09 | 48362268-48365696 | 898 | 101.5167 | 5.51 | nucl |
| GH_D09G2574.1 | 3 216 | D09 | 51563268-51567521 | 1071 | 119.46213 | 9.34 | nucl |
| GH_D10G1944.1 | 2 781 | D10 | 51528884-51532168 | 926 | 102.49744 | 8.92 | nucl |
| GH_D10G2421.1 | 2 397 | D10 | 61533814-61536564 | 798 | 91.18499 | 5.67 | chlo |
| GH_D11G0666.1 | 2 574 | D11 | 5396702-5399612 | 857 | 95.30735 | 9.44 | nucl |
| GH_D11G1341.1 | 2 355 | D11 | 12264012-12267490 | 784 | 88.40188 | 9.34 | nucl |
| GH_D11G1747.1 | 2 919 | D11 | 17898303-17902479 | 972 | 107.56187 | 8.73 | chlo |
| GH_D11G3080.1 | 2 610 | D11 | 61692025-61696072 | 869 | 95.91499 | 8.37 | nucl |
| GH_D11G3750.1 | 2 442 | D11 | 71069016-71072588 | 813 | 92.19382 | 5.86 | chlo |
| GH_D12G2464.1 | 1 251 | D12 | 57154546-57157210 | 416 | 46.98566 | 6.25 | nucl |
| GH_D12G2500.1 | 2 748 | D12 | 57445826-57454346 | 915 | 103.40408 | 8.21 | mito |
| GH_D12G2844.1 | 2 217 | D12 | 60412735-60415125 | 738 | 82.3026 | 9.26 | nucl |
| GH_D13G1252.1 | 2 736 | D13 | 39430684-39434564 | 911 | 101.67986 | 6.55 | nucl |
表1 陆地棉TRM基因家族理化性质
Tab.1 Physicochemical properties of Gossypium hirsutum L. TRM gene family members
| 基因ID Gene ID | 转录本长度 Transcript Length(bp) | 染色体 Chromo- some | 基因组位置 Genomic Location | 氨基酸数量 Number of Amino Acid | 分子量 Molecular Weight | 等电点 pI | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| GH_A01G1410.1 | 1 314 | A01 | 54808764-54811032 | 437 | 49.92686 | 4.88 | nucl |
| GH_A01G2253.1 | 1 518 | A01 | 115594835-115597597 | 505 | 57.79641 | 4.86 | nucl |
| GH_A01G2363.1 | 2 700 | A01 | 116832063-116835585 | 899 | 100.41775 | 6.11 | cyto |
| GH_A02G0234.1 | 2 757 | A02 | 2404926-2408330 | 918 | 103.50754 | 5.22 | nucl |
| GH_A02G2044.1 | 2 625 | A02 | 108074808-108078882 | 874 | 99.90307 | 6.04 | nucl |
| GH_A03G0533.1 | 1 563 | A03 | 8153312-8155305 | 520 | 60.28175 | 5.68 | nucl |
| GH_A03G1103.1 | 2 793 | A03 | 50408499-50412418 | 930 | 105.21887 | 5.07 | nucl |
| GH_A04G0979.1 | 1 434 | A04 | 69462180-69469536 | 477 | 52.62433 | 4.64 | nucl |
| GH_A05G0964.1 | 2 346 | A05 | 8607869-8610697 | 781 | 88.96019 | 6.01 | nucl |
| GH_A05G3931.1 | 3 123 | A05 | 102844278-102849879 | 1040 | 116.20696 | 9.31 | nucl |
| GH_A07G0059.1 | 1 533 | A07 | 628988-631260 | 510 | 58.37572 | 8.9 | nucl |
| GH_A07G0755.1 | 3 282 | A07 | 9022671-9027515 | 1093 | 120.99027 | 8.98 | nucl |
| GH_A07G2291.1 | 2 961 | D07 | 91905861-91909976 | 986 | 110.2077 | 6.3 | nucl |
| GH_A07G2350.1 | 2 331 | A07 | 92889383-92892178 | 776 | 87.42472 | 9.27 | nucl |
| GH_A08G0692.1 | 2 229 | A08 | 10127217-10130953 | 742 | 83.86942 | 9.19 | nucl |
| GH_A08G1551.1 | 1 131 | A08 | 102402502-102404142 | 376 | 42.99415 | 5.11 | nucl |
| GH_A08G2046.1 | 1 428 | A08 | 115647936-115650980 | 475 | 54.38497 | 5.52 | nucl |
| GH_A08G2091.1 | 1 770 | A08 | 116652133-116659356 | 589 | 67.4778 | 9.15 | chlo |
| GH_A08G2193.1 | 3 021 | A08 | 117970079-117975125 | 1006 | 112.14071 | 6.14 | nucl |
| GH_A09G0827.1 | 2 682 | A09 | 56330011-56333092 | 893 | 99.83975 | 9.11 | nucl |
| GH_A09G1541.1 | 1 524 | A09 | 71774616-71777153 | 507 | 57.473 | 5.23 | nucl |
| GH_A09G1756.1 | 2 982 | A09 | 74454345-74458907 | 993 | 110.9183 | 6.16 | nucl |
| GH_A09G1944.1 | 2 085 | A09 | 76570427-76572832 | 694 | 78.50671 | 9.56 | vacu |
| GH_A09G2222.1 | 2 697 | A09 | 79100072-79103507 | 898 | 101.38568 | 5.71 | chlo |
| GH_A09G2652.1 | 3 216 | A09 | 82737688-82741940 | 1071 | 119.47197 | 9.14 | nucl |
| GH_A10G1840.1 | 2 709 | A10 | 96909927-96913212 | 902 | 100.12998 | 9.1 | nucl |
| GH_A10G2325.1 | 2 406 | A10 | 109893241-109895998 | 801 | 91.39894 | 5.62 | nucl |
| GH_A11G0638.1 | 2 541 | A11 | 5673205-5676085 | 846 | 93.87066 | 9.49 | nucl |
| GH_A11G1309.1 | 2 355 | A11 | 13852756-13856249 | 784 | 88.58708 | 9.3 | nucl |
| GH_A11G1709.1 | 2 916 | A11 | 21197533-21201779 | 971 | 107.75294 | 8.55 | chlo |
| GH_A11G3051.1 | 2 604 | A11 | 111002265-111006157 | 867 | 96.23437 | 8.47 | nucl |
| GH_A11G3723.1 | 2 442 | A11 | 121125176-121128740 | 813 | 92.20752 | 5.62 | chlo |
| GH_A12G2452.1 | 1 251 | A12 | 102934223-102937012 | 416 | 47.26911 | 7.6 | nucl |
| GH_A12G2487.1 | 2 748 | A12 | 103276025-103279440 | 915 | 103.29828 | 8.32 | mito |
| GH_A12G2819.1 | 2 136 | A12 | 106188338-106190738 | 711 | 79.65164 | 9.26 | nucl |
| GH_A13G1327.1 | 2 727 | A13 | 78846901-78851063 | 908 | 101.54571 | 6.84 | nucl |
| GH_D01G1490.1 | 1 308 | D01 | 31939006-31941324 | 435 | 49.87765 | 4.87 | nucl |
| GH_D01G2334.1 | 1 515 | D01 | 62367964-62370708 | 504 | 57.81324 | 4.8 | nucl |
| GH_D01G2442.1 | 2 700 | D01 | 63514690-63518172 | 899 | 100.38982 | 5.99 | cyto |
| GH_D02G0253.1 | 2 760 | D02 | 2744183-2747593 | 919 | 103.89322 | 5.33 | mito |
| GH_D02G0972.1 | 1 272 | D02 | 20372804-20374278 | 423 | 48.19872 | 9.06 | nucl |
| GH_D02G1228.1 | 2 793 | D02 | 35813214-35817125 | 930 | 105.24384 | 4.93 | nucl |
| GH_D03G0017.1 | 2 625 | D03 | 138690-142736 | 874 | 99.91123 | 6.13 | nucl |
| GH_D03G1434.1 | 1 563 | D03 | 46317807-46319773 | 520 | 59.66096 | 5.74 | nucl |
| GH_D04G0442.1 | 3 126 | D04 | 6606757-6612330 | 1041 | 116.1618 | 9.36 | nucl |
| GH_D04G1311.1 | 2 853 | D04 | 43160828-43164823 | 950 | 105.2462 | 5.81 | nucl |
| GH_D05G0951.1 | 2 331 | D05 | 7672457-7675301 | 776 | 88.00605 | 6.03 | nucl |
| GH_D05G3052.1 | 2 688 | D05 | 31538189-31546485 | 895 | 102.41937 | 6.85 | nucl |
| GH_D07G0068.1 | 2 217 | D07 | 603139-606111 | 738 | 84.33982 | 9.02 | nucl |
| GH_D07G0751.1 | 3 279 | D07 | 8003392-8008211 | 1092 | 120.65422 | 9.27 | nucl |
| GH_D07G1883.1 | 2 877 | D07 | 38696495-38700181 | 958 | 109.3117 | 7.24 | nucl |
| GH_D07G2236.1 | 2 904 | D07 | 53803702-53807843 | 967 | 107.98503 | 5.97 | nucl |
| GH_D07G2293.1 | 1 923 | D07 | 54683888-54686682 | 640 | 72.50937 | 9.54 | nucl |
| GH_D08G0689.1 | 2 229 | D08 | 8659310-8663064 | 742 | 83.75555 | 9.36 | nucl |
| GH_D08G1569.1 | 1 131 | D08 | 51012741-51014358 | 376 | 42.8249 | 5.1 | nucl |
| GH_D08G2058.1 | 1 428 | D08 | 60460228-60462948 | 475 | 54.34586 | 5.45 | nucl |
| GH_D08G2107.1 | 1 725 | D08 | 61169567-61171897 | 574 | 65.66284 | 9.43 | chlo |
| GH_D08G2206.1 | 3 015 | D08 | 62410993-62416003 | 1004 | 112.23577 | 6.18 | nucl |
| GH_D09G0775.1 | 2 682 | D09 | 30764289-30767370 | 893 | 99.69663 | 9.03 | nucl |
| GH_D09G1524.1 | 1 548 | D09 | 42192153-42194698 | 515 | 58.35159 | 5 | nucl |
| GH_D09G1704.1 | 2 970 | D09 | 44087946-44092487 | 989 | 110.36687 | 6.26 | nucl |
| GH_D09G1898.1 | 2 070 | D09 | 46163346-46165731 | 689 | 77.82586 | 9.46 | vacu |
| GH_D09G2155.1 | 2 697 | D09 | 48362268-48365696 | 898 | 101.5167 | 5.51 | nucl |
| GH_D09G2574.1 | 3 216 | D09 | 51563268-51567521 | 1071 | 119.46213 | 9.34 | nucl |
| GH_D10G1944.1 | 2 781 | D10 | 51528884-51532168 | 926 | 102.49744 | 8.92 | nucl |
| GH_D10G2421.1 | 2 397 | D10 | 61533814-61536564 | 798 | 91.18499 | 5.67 | chlo |
| GH_D11G0666.1 | 2 574 | D11 | 5396702-5399612 | 857 | 95.30735 | 9.44 | nucl |
| GH_D11G1341.1 | 2 355 | D11 | 12264012-12267490 | 784 | 88.40188 | 9.34 | nucl |
| GH_D11G1747.1 | 2 919 | D11 | 17898303-17902479 | 972 | 107.56187 | 8.73 | chlo |
| GH_D11G3080.1 | 2 610 | D11 | 61692025-61696072 | 869 | 95.91499 | 8.37 | nucl |
| GH_D11G3750.1 | 2 442 | D11 | 71069016-71072588 | 813 | 92.19382 | 5.86 | chlo |
| GH_D12G2464.1 | 1 251 | D12 | 57154546-57157210 | 416 | 46.98566 | 6.25 | nucl |
| GH_D12G2500.1 | 2 748 | D12 | 57445826-57454346 | 915 | 103.40408 | 8.21 | mito |
| GH_D12G2844.1 | 2 217 | D12 | 60412735-60415125 | 738 | 82.3026 | 9.26 | nucl |
| GH_D13G1252.1 | 2 736 | D13 | 39430684-39434564 | 911 | 101.67986 | 6.55 | nucl |
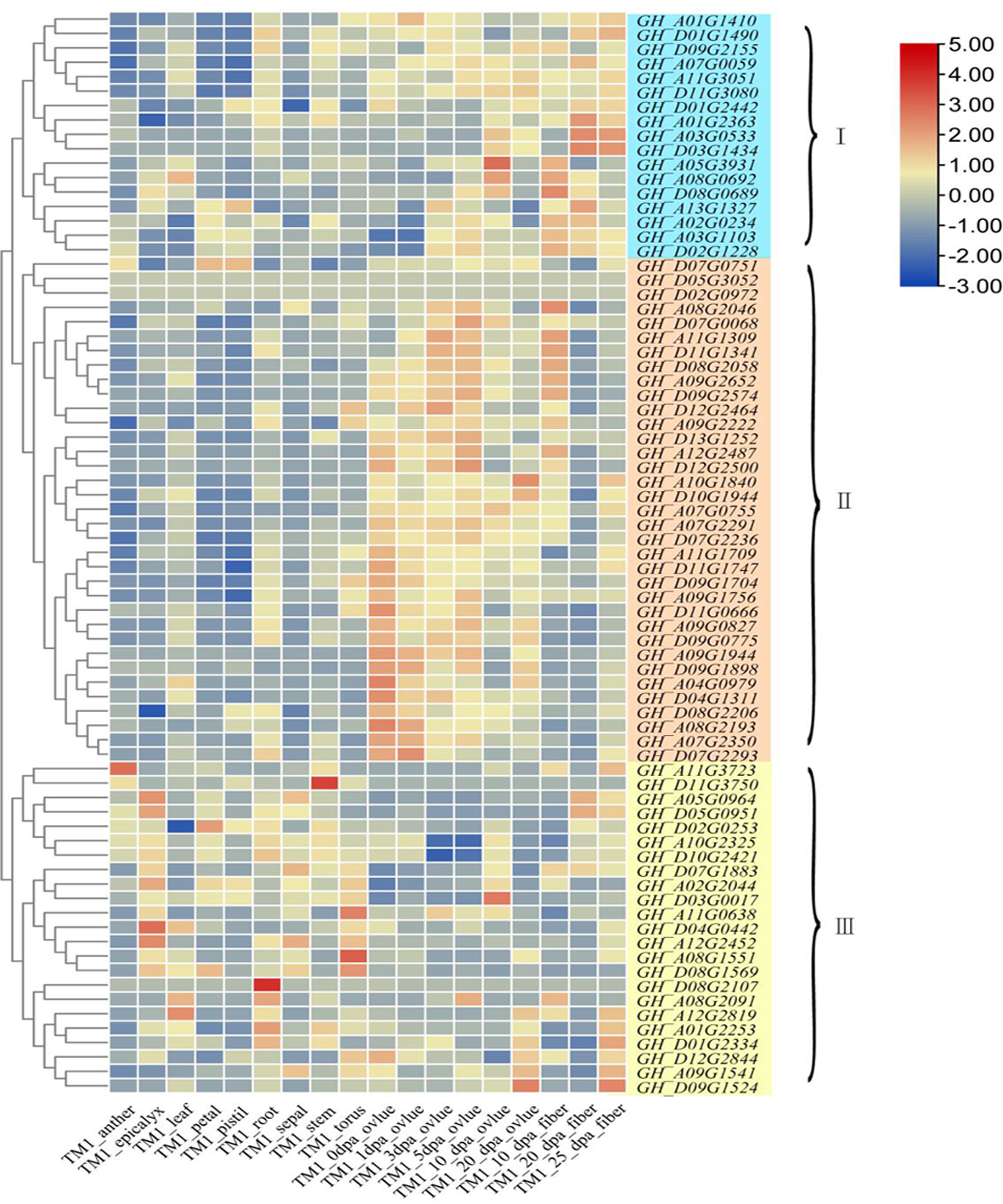
图7 陆地棉TRM基因组织表达热图 注(Note):花药:anther;萼状总苞:epicalyx;叶:leaf;花瓣:petal;雌蕊:pistil;根:root;花萼:sepal;茎:stem;花托:torus;胚珠:ovlue;纤维:fiber;开花后:dpa(d)
Fig.7 TRM gene expression heat map of Gossypium hirsutum L.
| 基因编号 Gene ID | 单倍型Haplotype | 单倍 型号 Hap | 纤维长度 FL P-value (t-test) | 纤维强度 FS P-value (t-test) | 马克隆值 FM P-value (t-test) |
|---|---|---|---|---|---|
| GH_A09G0827 | CT_A_C_T_G_C_T_C_A_A_A_C_C_T_T_T_A_C_G | Hap_0 | a | a | a |
| CT_A_C_T_G_C_T_C_A_A_A_C_C_T_T_T_A_C_G | Hap_1 | b | a | b | |
| C_A_T_T_A_C_T_C_A_C_A_T_C_T_A_T_A_C_A | Hap_2 | NA | NA | NA | |
| C_G_C_C_G_T_T_C_G_C_G_C_T_T_A_C_G_A_G | Hap_3 | a | b | ab | |
| C_A_T_T_A_C_T_C_A_C_A_T_C_T_A_T_A_C_G | Hap_4 | c | c | c | |
| GH_D09G0775 | G_C_T_G_GAAGCAC_C_T_C_T_G_A_G_G_C_C_G_C_A_A | Hap_0 | a | a | a |
| G_C_C_G_GAAGCAC_T_C_G_C_G_C_A_G_T_T_A_C_G_C | Hap_1 | b | b | b | |
| G_T_T_G_GAAGCAC_C_T_C_T_G_A_G_C_C_C_G_C_A_C | Hap_2 | a | ab | ab | |
| G_C_T_G_GAAGCAC_C_T_C_T_G_A_G_G_C_C_G_C_A_C | Hap_3 | a | ab | ab | |
| G_C_C_G_GAAGCAC_C_C_G_T_G_A_G_G_C_T_A_C_G_C | Hap_4 | c | c | ab | |
| GH_D09G1704 | CGTCGTG_A | Hap_0 | a | a | a |
| CGTCGTG_T | Hap_1 | b | b | b | |
| C_T | Hap_2 | ab | ab | ab | |
| GH_D11G0666 | C_G_C_GT_G_A_G_A_T_T_C_A_T_A_T_G_T_A_C | Hap_0 | a | a | a |
| T_G_C_GT_G_A_G_A_T_T_C_A_T_A_T_G_TA_A_C | Hap_1 | b | a | a | |
| C_A_CAA_G_A_T_A_T_C_G_C_G_C_C_C_G_T_A_T | Hap_2 | NA | NA | NA | |
| C_G_C_GT_G_A_G_A_T_T_C_A_T_A_T_G_TA_A_C | Hap_3 | ab | a | a | |
| GH_D08G2206 | G_C_T_A_A_T_G_G | Hap_0 | a | a | a |
| G_A_T_A_A_T_GA_G | Hap_1 | ab | b | a | |
| T_C_TC_C_T_TC_GA_C | Hap_2 | b | ab | b | |
| G_C_TC_C_T_TC_GA_C | Hap_3 | NA | NA | NA | |
| G_C_T_A_A_T_GA_G | Hap_4 | ab | ab | ab | |
| G_C_T_A_A_TC_GA_G | Hap_5 | NA | NA | NA | |
| GH_D10G1944 | G_A_GCA_AACT_T_T_C_G | Hap_0 | a | a | a |
| G_T_GCA_AACT_T_T_C_G | Hap_1 | b | b | a | |
| GH_A07G0755 | T_A_G_G_C_A_G_G_A_T_G_G_A_C_A_T_C_C_C_G_G_G_G_G_A_A_C | Hap_2 | a | a | a |
| T_A_C_G_C_A_G_G_A_T_G_G_A_C_A_T_C_C_C_G_G_G_G_G_A_A_C | Hap_3 | b | b | a | |
| A_A_C_G_C_A_C_C_T_C_G_G_G_CT_A_A_C_T_T_T_G_G_A_G_A_A_C | Hap_4 | c | c | b | |
| GH_A11G3051 | G_A_G_G_ACAG_C_G_T_C_G_C_G_A_G | Hap_0 | a | a | ab |
| G_A_G_G_ACAG_C_G_T_C_G_C_G_A_A | Hap_1 | b | a | a | |
| C_A_G_T_ACAG_A_G_T_C_T_A_A_A_G | Hap_2 | ab | a | b | |
| GH_D12G2844 | C_G_G_A_T_T_A_T_A | Hap_0 | a | a | a |
| C_G_G_A_T_T_A_G_A | Hap_1 | b | b | a | |
| C_G_T_G_A_T_T_G_G | Hap_2 | ab | ab | a | |
| GH_A09G1541 | C_G_A_T_A_A | Hap_0 | a | a | a |
| T_G_C_T_A_G | Hap_1 | b | b | b | |
| GH_D07G0751 | G_A_G_A_T_GT_C_A_T_CT_G_C_A_A_T_CA_G_A_A_AATATTAATT_CAT_C_T_A_C | Hap_0 | a | a | a |
| G_A_GAGTGGCAT_C_A_GT_C_G_T_CT_GA_A_A_A_T_CA_A_A_A_AATATTAATT_CAT_C_T_A_C | Hap_1 | b | ab | a | |
| G_A_GAGTGGCAT_C_A_GT_C_G_T_CT_GA_A_A_A_T_CA_A_A_A_AATATTAATT_CAT_C_C_A_C | Hap_2 | ab | b | b | |
| G_A_G_C_A_G_T_G_C_CT_G_A_A_G_T_CA_A_A_G_A_C_A_T_G_G | Hap_3 | abc | ab | ab | |
| G_G_G_C_A_GT_T_G_C_CT_G_A_A_G_T_CA_A_A_G_A_C_C_T_G_G | Hap_4 | NA | NA | NA | |
| C_G_G_C_A_GT_T_G_C_CT_G_A_A_G_T_CA_A_A_G_A_C_C_T_G_G | Hap_5 | c | ab | ab | |
| G_A_G_C_A_GT_C_G_T_C_G_A_G_A_C_C_A_T_A_AATATTAATT_CAT_C_T_A_C | Hap_6 | NA | NA | NA | |
| G_A_G_C_A_GT_C_G_T_CT_G_A_A_A_T_CA_A_A_A_AATATTAATT_CAT_C_T_A_C | Hap_7 | ab | ab | ab | |
| GH_A12G2487 | A_A_T_A_C_G_C_T_A_A_T_T_G_G_C_C_A | Hap_0 | a | a | a |
| A_C_T_T_C_G_T_C_A_A_C_C_A_A_C_C_A | Hap_1 | NA | NA | NA | |
| A_A_T_A_C_G_C_T_A_A_T_T_G_A_T_C_A | Hap_2 | a | a | a | |
| G_C_C_A_T_A_T_T_G_C_T_T_G_A_C_T_T | Hap_3 | a | a | b | |
| GH_D13G1252 | C_C_A_C_C | Hap_0 | a | a | a |
| C_G_A_C_G | Hap_1 | b | b | a | |
| GH_A07G2291 | T_T_C_C_T_G | Hap_0 | a | a | a |
| T_G_T_C_A_A | Hap_1 | b | b | a | |
| A_G_T_C_A_A | Hap_2 | c | c | b | |
| GH_D07G2236 | C_C_G_T_T_A_C_C_T | Hap_0 | a | a | ab |
| A_C_G_A_C_A_C_C_T | Hap_1 | b | b | a | |
| A_C_G_A_T_A_C_C_T | Hap_2 | ab | ab | b | |
| GH_D08G2058 | G_G_C_T_A_A_A_C_A_G_C_T_C_G | Hap_0 | a | a | a |
| G_T_T_A_C_T_A_T_T_C_C_T_C_G | Hap_1 | b | b | b | |
| G_T_T_A_C_T_A_T_A_C_A_A_C_G | Hap_2 | c | b | a | |
| G_T_T_A_C_T_A_T_A_C_C_T_C_G | Hap_3 | abc | b | ab | |
| GH_A08G2046 | T_G_A_C_G_C_A_C_G_G_G_A_T_T_C_C_T_G_T_A_G_TAATAACTAA_G_C_G_C_A_A_T_T_T_C_C_C_T_G_G_CG_T_C_C_T_A_T_T_T_T_C_AAATTAGGAAAAC_A_C_G_T_T_A_T_A_T_C_G_T_T_C | Hap_0 | a | a | a |
| T_G_A_G_G_T_A_C_G_G_G_A_T_C_C_G_C_G_C_G_G_TAATAACTAA_G_C_G_C_T_G_T_T_T_C_C_C_T_G_A_CG_G_C_C_T_G_A_T_T_T_C_A_G_C_G_T_T_A_T_A_T_C_G_T_T_C | Hap_1 | a | a | a | |
| C_G_A_G_G_C_G_A_G_A_A_C_C_C_T_C_C_G_C_A_G_TAATAACTAA_G_C_G_C_A_G_C_T_T_CT_C_C_C_G_A_CG_G_C_T_T_G_T_G_T_T_C_A_G_C_T_T_T_A_T_A_T_C_C_A_T_C | Hap_2 | a | a | a | |
| T_G_A_C_G_C_A_C_G_G_G_A_T_T_C_C_T_G_T_A_G_TAATAACTAA_G_C_G_C_A_A_T_T_T_C_C_C_T_G_G_CG_T_C_C_T_A_T_T_T_T_C_A_A_C_G_T_T_A_T_A_T_C_G_T_T_C | Hap_3 | a | a | a | |
| T_G_A_G_G_C_A_C_A_G_G_A_T_C_C_C_C_T_C_A_G_TAATAACTAA_G_C_T_C_A_G_T_T_T_C_T_C_T_G_A_CG_G_C_C_T_G_T_T_T_G_C_A_G_C_G_T_T_A_T_A_T_T_G_T_T_C | Hap_4 | NA | NA | NA | |
| T_G_A_C_G_C_A_C_G_G_G_A_T_C_C_C_C_G_T_A_G_TAATAACTAA_G_C_G_C_A_G_T_T_T_C_C_C_T_G_G_CG_T_C_C_T_A_T_T_T_T_C_AAATTAGGAAAAC_A_C_G_T_T_A_T_A_T_C_G_T_T_C | Hap_5 | a | a | b | |
| GH_D09G2574 | C_C_T_C_G_C_T_T_G_C_C_G_C_A_G_C_T_A_C_A_C_T | Hap_0 | a | a | a |
| T_C_T_C_G_C_T_C_G_C_C_G_C_G_G_C_T_A_T_A_C_T | Hap_1 | a | a | b | |
| C_C_C_T_A_T_C_C_A_T_T_G_C_G_A_T_G_C_C_C_T_T | Hap_2 | b | b | ab | |
| C_T_T_C_G_C_T_C_G_C_C_G_C_A_G_C_T_A_C_A_C_C | Hap_3 | a | a | ab | |
| GH_D11G1341 | C_C_G_C_C_C_G_T_A_C_CA | Hap_0 | a | a | a |
| A_C_G_C_A_C_G_T_G_C_CA | Hap_1 | b | b | a | |
| GH_D12G2464 | C_C_C_G_G_A_A_C_A_A_G_T_A | Hap_0 | a | a | a |
| T_A_C_G_G_A_C_C_A_A_G_T_C | Hap_1 | b | b | b | |
| C_C_T_A_G_A_C_T_A_T_T_A_A | Hap_2 | a | a | a | |
| GH_A09G2222 | T_A_C_T_T_C_A_AATTTTAAAATTT_T_TA_G_C_G_A_A_TC_G_C_GA_A_C | Hap_0 | a | a | a |
| T_A_C_C_T_C_A_AATTTTAAAATTT_TA_T_A_C_G_G_A_T_G_C_G_A_C | Hap_1 | b | b | a | |
| GH_D07G0068 | A_G_G_C_C_G_C_C_A_C_T_T_C_T_C | Hap_0 | a | a | a |
| A_A_GA_C_T_A_T_C_A_T_C_C_C_A_T | Hap_1 | b | b | a | |
| GH_D08G0689 | G_T_C_C_A_A_A_A_C_C_T_C_C_A_G_T_G_C_G_G_T_T_G_G_CA_A_C_C_G_A_C_G_A_C | Hap_0 | a | a | a |
| G_C_C_C_A_A_A_A_C_C_T_C_C_A_G_C_G_C_G_G_A_T_G_G_CA_A_C_C_G_A_C_T_A_T | Hap_1 | b | b | b | |
| GH_A05G3931 | T_T_A_G_CCGACA_C_C_T | Hap_0 | a | a | a |
| T_C_G_G_C_A_C_T | Hap_1 | b | b | a | |
| G_C_G_A_CCGACA_A_T_A | Hap_2 | abc | bc | a | |
| T_C_A_G_CCGACA_A_C_T | Hap_3 | c | c | a | |
| GH_A13G1327 | G_A_T_G_G_T_A_C_A_C_G_C_T_C_T | Hap_0 | a | a | a |
| G_G_G_C_G_T_C_C_G_C_A_C_A_C_T | Hap_1 | b | b | b | |
| G_A_T_G_G_C_A_C_A_C_G_C_T_C_T | Hap_2 | b | b | c | |
| G_A_T_C_G_T_A_T_A_C_G_C_A_A_A | Hap_3 | c | c | d | |
| GH_A01G1410 | A_T_C_A_A_G_A_G_C_A_C_C_A_A_T_T | Hap_0 | a | a | a |
| T_C_C_A_G_G_A_G_C_A_T_C_A_G_T_C | Hap_1 | a | a | b | |
| T_C_C_A_G_G_A_G_C_A_T_C_G_G_T_C | Hap_2 | b | a | b | |
| GH_A01G2363 | T_A_C_C_G_G_A_C_T_G_C_A_A_A_C | Hap_0 | a | a | a |
| T_G_T_T_G_A_A_C_T_G_T_G_G_A_A | Hap_1 | ab | ab | ab | |
| T_G_T_T_G_G_A_C_T_G_C_G_A_A_C | Hap_2 | a | ab | b | |
| T_G_T_T_A_G_A_T_T_A_C_G_A_G_C | Hap_3 | b | b | ab | |
| GH_D03G1434 | C_T_G_C_A_A_C_A_G_A_T_C_G_G | Hap_0 | a | a | a |
| T_C_GTT_CT_T_A_T_A_G_A_T_C_G_T | Hap_1 | a | b | b | |
| T_T_G_C_A_A_T_A_C_A_A_C_G_G | Hap_2 | b | c | b | |
| T_C_GTT_CT_T_A_T_A_G_A_T_C_G_G | Hap_3 | c | d | c | |
| GH_A11G3723 | A_C_C_A_G_C_C_T_G_A_G_A_C_AC_A | Hap_0 | a | a | a |
| A_C_C_G_G_C_C_T_A_A_C_A_C_AC_C | Hap_1 | a | a | a | |
| T_C_C_G_A_C_C_T_G_T_C_A_C_A_C | Hap_2 | b | b | b | |
| A_G_A_G_G_C_C_T_G_A_C_G_T_AC_C | Hap_3 | NA | NA | NA | |
| A_C_C_G_G_C_C_T_G_A_C_A_C_AC_C | Hap_4 | NA | NA | NA | |
| A_C_C_G_G_A_T_G_G_A_C_A_C_AC_C | Hap_5 | NA | NA | NA | |
| GH_D02G0972 | A_C_A_C_T_T_C_C_G_G_G | Hap_0 | a | a | a |
| A_C_A_C_T_T_C_C_G_A_G | Hap_1 | b | b | b | |
| T_T_G_A_T_T_A_C_A_G_T | Hap_2 | NA | NA | NA | |
| GH_A10G2325 | A_T_G_A_G_C_T | Hap_0 | a | a | a |
| A_T_G_A_G_C_C | Hap_1 | b | b | b | |
| T_C_A_T_T_T_C | Hap_2 | ab | c | b | |
| GH_D10G2421 | C_T_T_A_G_T_A_G_C_A_T_C_A_T_A_G_A_T_C_G_C_A_A_C_A_A_TC_A_T_C_C_T_G_G_T_A_T_A_G_A_A_G_G_TG_C_A_T_C_TCACTCAATGTGCTAATATGTCACATAACAATCCATTAAAAG AAAACAAAAAA_T_T_T_G_C_T_G_T_T_A_T_A_T_T_G_T_A_G_GA_C_C_G_G_G | Hap_0 | a | a | a |
| C_G_T_A_T_G_C_T_T_G_C_A_G_C_G_A_T_G_A_A_C_A_G_G_G_G_T_G_C_G_A_C_A_A_A_G_G_T_C_G_A_G_G_T_T_G_T_T_T_T_T_C_GTAC_T_A_A_C_G_G_C_A_A_G_A_C_T_C_G_A_G_G_G_G | Hap_1 | b | b | b | |
| C_T_T_A_G_T_A_G_C_A_T_C_A_C_A_G_A_T_C_G_C_A_A_C_A_A_TC_A_T_C_C_T_G_G_T_A_T_A_G_A_A_G_G_TG_C_A_T_C_TCACTCAATGTGCTAATATGTCACATAACAATCCATTAAAAGAAAA CAAAAAA_T_A_T_G_C_T_G_T_T_A_T_G_T_T_G_T_A_G_GA_C_C_G_G_A | Hap_2 | ab | ab | ab | |
| T_G_T_A_G_T_A_G_C_A_T_A_A_C_A_G_A_T_C_G_C_A_A_C_A_A_TC_A_T_C_C_T_G_G_T_A_T_A_G_A_A_G_G_TG_C_A_T_C_TCACTCAATGTGCTAATATGTCACATAACAATCCATTAAAAGAAAAC AAAAAA_T_T_T_G_C_T_G_T_T_A_T_A_T_T_G_T_A_G_GA_C_C_G_G_G | Hap_3 | NA | NA | NA | |
| C_T_T_A_G_T_A_G_C_A_T_C_A_C_A_G_A_T_C_G_C_A_A_C_A_A_TC_A_T_C_C_T_G_G_T_A_T_A_G_A_A_G_G_TG_C_A_T_C_TCACTCAATGTGCTAATATGTCACATAACAATCCATTAAAAGAAAAC AAAAAA_C_T_T_G_C_T_G_T_T_A_T_A_T_T_G_T_A_G_GA_C_C_G_G_A | Hap_4 | NA | NA | NA | |
| C_T_T_A_G_T_A_G_C_A_T_C_A_C_A_G_A_T_C_G_C_A_A_C_A_A_TC_A_T_C_C_T_G_G_T_A_T_A_G_A_A_G_G_TG_C_A_T_C_TCACTCAATGTGCTAATATGTCACATAACAATCCATTAAAAGAAAA CAAAAAA_T_T_T_G_C_T_G_T_T_A_T_A_T_T_G_T_A_G_GA_C_C_G_G_A | Hap_5 | NA | NA | NA | |
| GH_A12G2819 | G_A_G_G_C_C_A_G_T_G_CAGTGGGGACGAAGTT | Hap_0 | a | a | a |
| G_A_G_G_C_C_A_T_T_G_CAGTGGGGACGAAGTT | Hap_1 | b | b | b | |
| A_C_G_C_T_G_C_T_C_T_C | Hap_2 | NA | NA | NA | |
| G_C_T_G_C_G_A_T_C_G_CAGTGGGGACGAAGTT | Hap_3 | NA | NA | NA | |
| GH_A01G2253 | A_C_G_G_A_C_A_A_A_G_T_A_G_T_A_C | Hap_0 | a | a | a |
| A_T_G_A_C_C_T_A_G_C_TAA_A_A_C_A_C | Hap_1 | a | a | b | |
| C_C_G_A_A_G_T_A_G_C_T_T_A_C_A_A | Hap_2 | a | a | ab | |
| C_C_C_A_A_G_T_A_G_C_T_T_A_C_G_C | Hap_3 | a | a | ab | |
| A_C_G_A_A_C_A_T_G_C_T_A_A_C_A_C | Hap_4 | a | a | NA | |
| A_C_G_A_A_C_A_A_G_C_T_A_A_T_A_C | Hap_5 | a | a | NA | |
| GH_D08G2107 | C_G_T_G_A_C_A_G_A_A_T_G_T_T_A_A | Hap_0 | a | a | a |
| G_T_C_C_T_C_G_T_G_C_T_G_C_G_A_A | Hap_1 | b | b | b | |
| G_T_C_C_A_G_A_G_A_C_C_C_C_G_G_G | Hap_2 | NA | NA | NA | |
| G_T_C_C_A_G_A_G_A_C_T_C_C_G_G_G | Hap_3 | c | ab | ab | |
| GH_A08G2091 | A_A_T_C_C_G_C_T_C_G_C_T_AG_G_T_G_G_T_G_G_A | Hap_0 | a | a | a |
| A_A_T_C_C_G_T_T_C_G_C_T_AG_G_T_G_G_T_G_G_A | Hap_1 | b | b | a | |
| A_T_T_T_C_A_C_A_C_G_C_A_A_A_A_C_T_A_A_G_A | Hap_2 | ab | a | a | |
| A_A_T_C_T_A_C_T_C_G_C_T_A_A_A_C_T_A_A_G_A | Hap_3 | NA | NA | a | |
| A_A_T_T_C_A_C_T_C_G_C_A_A_A_A_C_T_A_A_G_A | Hap_4 | NA | NA | a | |
| A_A_T_C_C_G_C_T_C_G_C_T_AG_G_T_G_G_T_G_A_A | Hap_5 | ab | a | a | |
| C_A_A_C_C_A_C_T_T_T_A_T_AG_A_A_G_G_T_A_G_G | Hap_6 | ab | ab | a | |
| GH_D09G2155 | C_G_T_G | Hap_0 | a | a | a |
| A_T_T_G | Hap_1 | a | b | b | |
| C_G_T_T | Hap_2 | b | c | ab | |
| C_G_C_G | Hap_3 | NA | NA | NA | |
| A_G_T_G | Hap_4 | NA | NA | NA | |
| GH_D04G0442 | A_G_G_G_C_A_T_T_T | Hap_0 | a | a | a |
| A_G_A_G_C_G_T_C_T | Hap_1 | b | b | b | |
| T_A_G_G_G_G_T_T_G | Hap_2 | NA | NA | NA | |
| A_G_G_A_C_G_G_T_T | Hap_3 | NA | NA | NA | |
| A_G_G_G_C_G_T_T_T | Hap_4 | ab | ab | ab | |
| GH_A05G0964 | T_A_G_G_A_C_T_C_C_A_A_A_A_A_A_G | Hap_0 | a | a | a |
| T_A_G_G_T_C_C_C_C_A_A_A_A_A_A_G | Hap_1 | b | b | b | |
| A_A_G_T_A_T_T_A_T_T_T_A_T_C_AT_T | Hap_2 | NA | NA | NA | |
| T_G_T_G_A_C_T_C_C_A_A_T_A_A_A_G | Hap_3 | NA | NA | NA | |
| GH_D05G0951 | G_T_A_G | Hap_0 | a | a | a |
| A_C_G_T | Hap_1 | a | a | b | |
| G_T_A_T | Hap_2 | NA | NA | NA | |
| GH_A11G0638 | A_T_G_C_T_G_C_T_G_C_A_C_C_A_C | Hap_0 | a | a | a |
| G_T_A_G_C_A_C_A_G_T_A_A_C_G_C | Hap_1 | a | a | b | |
| A_A_G_G_T_G_C_T_A_C_C_C_G_A_T | Hap_2 | a | ab | ab | |
| G_T_A_G_C_G_C_A_G_T_A_A_C_G_C | Hap_3 | b | b | a | |
| A_A_G_G_T_G_T_T_A_C_C_C_G_A_T | Hap_4 | NA | NA | NA | |
| A_T_G_G_T_G_C_T_G_C_A_C_C_A_C | Hap_5 | a | ab | ab | |
| G_T_G_G_C_G_C_A_G_C_A_A_C_G_C | Hap_6 | b | ab | ab | |
| A_T_G_G_T_G_C_T_A_C_C_C_C_A_T | Hap_7 | NA | NA | NA | |
| GH_A08G1551 | T_A_C_G_G_T_C_T_A_T_C_T_G_T | Hap_0 | a | a | a |
| G_AT_C_A_A_G_A_C_G_G_T_C_C_G | Hap_1 | NA | NA | a | |
| T_A_A_G_G_T_C_T_A_T_C_T_G_T | Hap_2 | b | b | a | |
| GH_D08G1569 | T_A_T_G_C | Hap_0 | a | a | a |
| T_G_C_G_T | Hap_1 | b | a | a | |
| T_A_C_G_T | Hap_2 | NA | NA | NA | |
| A_A_T_T_C | Hap_3 | NA | NA | NA | |
| GH_D07G1883 | G_TAGA_G_C_T_G_T_C_T | Hap_0 | a | a | a |
| A_TAGA_G_T_A_A_C_A_T | Hap_1 | NA | NA | NA | |
| A_T_A_T_A_A_C_A_T | Hap_2 | a | b | b | |
| G_TAGA_G_C_T_G_T_C_C | Hap_3 | NA | NA | NA | |
| G_TAGA_G_C_T_A_T_C_T | Hap_4 | NA | NA | NA | |
| GH_A12G2452 | T_A_C_T_G_C_C_T_T_T_CT_T_C_G_TAAATTTTCTTAATTTCCGTTATTCAACCACAATATTTTTTAA_T | Hap_0 | a | a | a |
| C_G_C_C_G_C_C_C_T_T_C_T_C_A_TAAATTTTCTTAATTTCCGTTATTCAACCACAATATTTTTTAA_T | Hap_1 | a | b | b | |
| T_A_A_T_G_T_T_T_T_A_C_A_T_A_T_TTCAAAACGTTTGATGTAACCACTAA | Hap_2 | NA | NA | NA | |
| T_A_C_T_A_C_C_T_T_T_C_T_C_A_TAAATTTTCTTAATTTCCGTTATTCAACCACAATATTTTTTAA_T | Hap_3 | b | c | ab | |
| T_A_C_T_A_C_C_T_C_T_C_T_C_A_TAAATTTTCTTAATTTCCGTTATTCAACCACAATATTTTTTAA_T | Hap_4 | NA | NA | NA | |
| GH_A02G2044 | C_G_A_A_A_G_G_C_G_G_C_A_G_C_G_G_G | Hap_0 | a | a | a |
| C_G_A_G_A_G_G_C_G_G_C_A_G_T_G_G_G | Hap_1 | b | b | b | |
| T_G_G_G_G_C_G_T_T_C_T_G_A_C_G_G_A | Hap_2 | ab | ab | ab | |
| C_G_G_G_A_G_C_C_G_C_C_G_G_C_C_A_A | Hap_3 | ab | b | ab | |
| C_A_A_G_A_G_G_C_G_G_C_A_G_C_G_G_G | Hap_4 | NA | NA | NA |
表2 陆地棉TRM基因家族成员群体中单倍型的性状差异
Tab.2 Character differences of haplotypes among TRM gene family members in Gossypium hirsutum L.
| 基因编号 Gene ID | 单倍型Haplotype | 单倍 型号 Hap | 纤维长度 FL P-value (t-test) | 纤维强度 FS P-value (t-test) | 马克隆值 FM P-value (t-test) |
|---|---|---|---|---|---|
| GH_A09G0827 | CT_A_C_T_G_C_T_C_A_A_A_C_C_T_T_T_A_C_G | Hap_0 | a | a | a |
| CT_A_C_T_G_C_T_C_A_A_A_C_C_T_T_T_A_C_G | Hap_1 | b | a | b | |
| C_A_T_T_A_C_T_C_A_C_A_T_C_T_A_T_A_C_A | Hap_2 | NA | NA | NA | |
| C_G_C_C_G_T_T_C_G_C_G_C_T_T_A_C_G_A_G | Hap_3 | a | b | ab | |
| C_A_T_T_A_C_T_C_A_C_A_T_C_T_A_T_A_C_G | Hap_4 | c | c | c | |
| GH_D09G0775 | G_C_T_G_GAAGCAC_C_T_C_T_G_A_G_G_C_C_G_C_A_A | Hap_0 | a | a | a |
| G_C_C_G_GAAGCAC_T_C_G_C_G_C_A_G_T_T_A_C_G_C | Hap_1 | b | b | b | |
| G_T_T_G_GAAGCAC_C_T_C_T_G_A_G_C_C_C_G_C_A_C | Hap_2 | a | ab | ab | |
| G_C_T_G_GAAGCAC_C_T_C_T_G_A_G_G_C_C_G_C_A_C | Hap_3 | a | ab | ab | |
| G_C_C_G_GAAGCAC_C_C_G_T_G_A_G_G_C_T_A_C_G_C | Hap_4 | c | c | ab | |
| GH_D09G1704 | CGTCGTG_A | Hap_0 | a | a | a |
| CGTCGTG_T | Hap_1 | b | b | b | |
| C_T | Hap_2 | ab | ab | ab | |
| GH_D11G0666 | C_G_C_GT_G_A_G_A_T_T_C_A_T_A_T_G_T_A_C | Hap_0 | a | a | a |
| T_G_C_GT_G_A_G_A_T_T_C_A_T_A_T_G_TA_A_C | Hap_1 | b | a | a | |
| C_A_CAA_G_A_T_A_T_C_G_C_G_C_C_C_G_T_A_T | Hap_2 | NA | NA | NA | |
| C_G_C_GT_G_A_G_A_T_T_C_A_T_A_T_G_TA_A_C | Hap_3 | ab | a | a | |
| GH_D08G2206 | G_C_T_A_A_T_G_G | Hap_0 | a | a | a |
| G_A_T_A_A_T_GA_G | Hap_1 | ab | b | a | |
| T_C_TC_C_T_TC_GA_C | Hap_2 | b | ab | b | |
| G_C_TC_C_T_TC_GA_C | Hap_3 | NA | NA | NA | |
| G_C_T_A_A_T_GA_G | Hap_4 | ab | ab | ab | |
| G_C_T_A_A_TC_GA_G | Hap_5 | NA | NA | NA | |
| GH_D10G1944 | G_A_GCA_AACT_T_T_C_G | Hap_0 | a | a | a |
| G_T_GCA_AACT_T_T_C_G | Hap_1 | b | b | a | |
| GH_A07G0755 | T_A_G_G_C_A_G_G_A_T_G_G_A_C_A_T_C_C_C_G_G_G_G_G_A_A_C | Hap_2 | a | a | a |
| T_A_C_G_C_A_G_G_A_T_G_G_A_C_A_T_C_C_C_G_G_G_G_G_A_A_C | Hap_3 | b | b | a | |
| A_A_C_G_C_A_C_C_T_C_G_G_G_CT_A_A_C_T_T_T_G_G_A_G_A_A_C | Hap_4 | c | c | b | |
| GH_A11G3051 | G_A_G_G_ACAG_C_G_T_C_G_C_G_A_G | Hap_0 | a | a | ab |
| G_A_G_G_ACAG_C_G_T_C_G_C_G_A_A | Hap_1 | b | a | a | |
| C_A_G_T_ACAG_A_G_T_C_T_A_A_A_G | Hap_2 | ab | a | b | |
| GH_D12G2844 | C_G_G_A_T_T_A_T_A | Hap_0 | a | a | a |
| C_G_G_A_T_T_A_G_A | Hap_1 | b | b | a | |
| C_G_T_G_A_T_T_G_G | Hap_2 | ab | ab | a | |
| GH_A09G1541 | C_G_A_T_A_A | Hap_0 | a | a | a |
| T_G_C_T_A_G | Hap_1 | b | b | b | |
| GH_D07G0751 | G_A_G_A_T_GT_C_A_T_CT_G_C_A_A_T_CA_G_A_A_AATATTAATT_CAT_C_T_A_C | Hap_0 | a | a | a |
| G_A_GAGTGGCAT_C_A_GT_C_G_T_CT_GA_A_A_A_T_CA_A_A_A_AATATTAATT_CAT_C_T_A_C | Hap_1 | b | ab | a | |
| G_A_GAGTGGCAT_C_A_GT_C_G_T_CT_GA_A_A_A_T_CA_A_A_A_AATATTAATT_CAT_C_C_A_C | Hap_2 | ab | b | b | |
| G_A_G_C_A_G_T_G_C_CT_G_A_A_G_T_CA_A_A_G_A_C_A_T_G_G | Hap_3 | abc | ab | ab | |
| G_G_G_C_A_GT_T_G_C_CT_G_A_A_G_T_CA_A_A_G_A_C_C_T_G_G | Hap_4 | NA | NA | NA | |
| C_G_G_C_A_GT_T_G_C_CT_G_A_A_G_T_CA_A_A_G_A_C_C_T_G_G | Hap_5 | c | ab | ab | |
| G_A_G_C_A_GT_C_G_T_C_G_A_G_A_C_C_A_T_A_AATATTAATT_CAT_C_T_A_C | Hap_6 | NA | NA | NA | |
| G_A_G_C_A_GT_C_G_T_CT_G_A_A_A_T_CA_A_A_A_AATATTAATT_CAT_C_T_A_C | Hap_7 | ab | ab | ab | |
| GH_A12G2487 | A_A_T_A_C_G_C_T_A_A_T_T_G_G_C_C_A | Hap_0 | a | a | a |
| A_C_T_T_C_G_T_C_A_A_C_C_A_A_C_C_A | Hap_1 | NA | NA | NA | |
| A_A_T_A_C_G_C_T_A_A_T_T_G_A_T_C_A | Hap_2 | a | a | a | |
| G_C_C_A_T_A_T_T_G_C_T_T_G_A_C_T_T | Hap_3 | a | a | b | |
| GH_D13G1252 | C_C_A_C_C | Hap_0 | a | a | a |
| C_G_A_C_G | Hap_1 | b | b | a | |
| GH_A07G2291 | T_T_C_C_T_G | Hap_0 | a | a | a |
| T_G_T_C_A_A | Hap_1 | b | b | a | |
| A_G_T_C_A_A | Hap_2 | c | c | b | |
| GH_D07G2236 | C_C_G_T_T_A_C_C_T | Hap_0 | a | a | ab |
| A_C_G_A_C_A_C_C_T | Hap_1 | b | b | a | |
| A_C_G_A_T_A_C_C_T | Hap_2 | ab | ab | b | |
| GH_D08G2058 | G_G_C_T_A_A_A_C_A_G_C_T_C_G | Hap_0 | a | a | a |
| G_T_T_A_C_T_A_T_T_C_C_T_C_G | Hap_1 | b | b | b | |
| G_T_T_A_C_T_A_T_A_C_A_A_C_G | Hap_2 | c | b | a | |
| G_T_T_A_C_T_A_T_A_C_C_T_C_G | Hap_3 | abc | b | ab | |
| GH_A08G2046 | T_G_A_C_G_C_A_C_G_G_G_A_T_T_C_C_T_G_T_A_G_TAATAACTAA_G_C_G_C_A_A_T_T_T_C_C_C_T_G_G_CG_T_C_C_T_A_T_T_T_T_C_AAATTAGGAAAAC_A_C_G_T_T_A_T_A_T_C_G_T_T_C | Hap_0 | a | a | a |
| T_G_A_G_G_T_A_C_G_G_G_A_T_C_C_G_C_G_C_G_G_TAATAACTAA_G_C_G_C_T_G_T_T_T_C_C_C_T_G_A_CG_G_C_C_T_G_A_T_T_T_C_A_G_C_G_T_T_A_T_A_T_C_G_T_T_C | Hap_1 | a | a | a | |
| C_G_A_G_G_C_G_A_G_A_A_C_C_C_T_C_C_G_C_A_G_TAATAACTAA_G_C_G_C_A_G_C_T_T_CT_C_C_C_G_A_CG_G_C_T_T_G_T_G_T_T_C_A_G_C_T_T_T_A_T_A_T_C_C_A_T_C | Hap_2 | a | a | a | |
| T_G_A_C_G_C_A_C_G_G_G_A_T_T_C_C_T_G_T_A_G_TAATAACTAA_G_C_G_C_A_A_T_T_T_C_C_C_T_G_G_CG_T_C_C_T_A_T_T_T_T_C_A_A_C_G_T_T_A_T_A_T_C_G_T_T_C | Hap_3 | a | a | a | |
| T_G_A_G_G_C_A_C_A_G_G_A_T_C_C_C_C_T_C_A_G_TAATAACTAA_G_C_T_C_A_G_T_T_T_C_T_C_T_G_A_CG_G_C_C_T_G_T_T_T_G_C_A_G_C_G_T_T_A_T_A_T_T_G_T_T_C | Hap_4 | NA | NA | NA | |
| T_G_A_C_G_C_A_C_G_G_G_A_T_C_C_C_C_G_T_A_G_TAATAACTAA_G_C_G_C_A_G_T_T_T_C_C_C_T_G_G_CG_T_C_C_T_A_T_T_T_T_C_AAATTAGGAAAAC_A_C_G_T_T_A_T_A_T_C_G_T_T_C | Hap_5 | a | a | b | |
| GH_D09G2574 | C_C_T_C_G_C_T_T_G_C_C_G_C_A_G_C_T_A_C_A_C_T | Hap_0 | a | a | a |
| T_C_T_C_G_C_T_C_G_C_C_G_C_G_G_C_T_A_T_A_C_T | Hap_1 | a | a | b | |
| C_C_C_T_A_T_C_C_A_T_T_G_C_G_A_T_G_C_C_C_T_T | Hap_2 | b | b | ab | |
| C_T_T_C_G_C_T_C_G_C_C_G_C_A_G_C_T_A_C_A_C_C | Hap_3 | a | a | ab | |
| GH_D11G1341 | C_C_G_C_C_C_G_T_A_C_CA | Hap_0 | a | a | a |
| A_C_G_C_A_C_G_T_G_C_CA | Hap_1 | b | b | a | |
| GH_D12G2464 | C_C_C_G_G_A_A_C_A_A_G_T_A | Hap_0 | a | a | a |
| T_A_C_G_G_A_C_C_A_A_G_T_C | Hap_1 | b | b | b | |
| C_C_T_A_G_A_C_T_A_T_T_A_A | Hap_2 | a | a | a | |
| GH_A09G2222 | T_A_C_T_T_C_A_AATTTTAAAATTT_T_TA_G_C_G_A_A_TC_G_C_GA_A_C | Hap_0 | a | a | a |
| T_A_C_C_T_C_A_AATTTTAAAATTT_TA_T_A_C_G_G_A_T_G_C_G_A_C | Hap_1 | b | b | a | |
| GH_D07G0068 | A_G_G_C_C_G_C_C_A_C_T_T_C_T_C | Hap_0 | a | a | a |
| A_A_GA_C_T_A_T_C_A_T_C_C_C_A_T | Hap_1 | b | b | a | |
| GH_D08G0689 | G_T_C_C_A_A_A_A_C_C_T_C_C_A_G_T_G_C_G_G_T_T_G_G_CA_A_C_C_G_A_C_G_A_C | Hap_0 | a | a | a |
| G_C_C_C_A_A_A_A_C_C_T_C_C_A_G_C_G_C_G_G_A_T_G_G_CA_A_C_C_G_A_C_T_A_T | Hap_1 | b | b | b | |
| GH_A05G3931 | T_T_A_G_CCGACA_C_C_T | Hap_0 | a | a | a |
| T_C_G_G_C_A_C_T | Hap_1 | b | b | a | |
| G_C_G_A_CCGACA_A_T_A | Hap_2 | abc | bc | a | |
| T_C_A_G_CCGACA_A_C_T | Hap_3 | c | c | a | |
| GH_A13G1327 | G_A_T_G_G_T_A_C_A_C_G_C_T_C_T | Hap_0 | a | a | a |
| G_G_G_C_G_T_C_C_G_C_A_C_A_C_T | Hap_1 | b | b | b | |
| G_A_T_G_G_C_A_C_A_C_G_C_T_C_T | Hap_2 | b | b | c | |
| G_A_T_C_G_T_A_T_A_C_G_C_A_A_A | Hap_3 | c | c | d | |
| GH_A01G1410 | A_T_C_A_A_G_A_G_C_A_C_C_A_A_T_T | Hap_0 | a | a | a |
| T_C_C_A_G_G_A_G_C_A_T_C_A_G_T_C | Hap_1 | a | a | b | |
| T_C_C_A_G_G_A_G_C_A_T_C_G_G_T_C | Hap_2 | b | a | b | |
| GH_A01G2363 | T_A_C_C_G_G_A_C_T_G_C_A_A_A_C | Hap_0 | a | a | a |
| T_G_T_T_G_A_A_C_T_G_T_G_G_A_A | Hap_1 | ab | ab | ab | |
| T_G_T_T_G_G_A_C_T_G_C_G_A_A_C | Hap_2 | a | ab | b | |
| T_G_T_T_A_G_A_T_T_A_C_G_A_G_C | Hap_3 | b | b | ab | |
| GH_D03G1434 | C_T_G_C_A_A_C_A_G_A_T_C_G_G | Hap_0 | a | a | a |
| T_C_GTT_CT_T_A_T_A_G_A_T_C_G_T | Hap_1 | a | b | b | |
| T_T_G_C_A_A_T_A_C_A_A_C_G_G | Hap_2 | b | c | b | |
| T_C_GTT_CT_T_A_T_A_G_A_T_C_G_G | Hap_3 | c | d | c | |
| GH_A11G3723 | A_C_C_A_G_C_C_T_G_A_G_A_C_AC_A | Hap_0 | a | a | a |
| A_C_C_G_G_C_C_T_A_A_C_A_C_AC_C | Hap_1 | a | a | a | |
| T_C_C_G_A_C_C_T_G_T_C_A_C_A_C | Hap_2 | b | b | b | |
| A_G_A_G_G_C_C_T_G_A_C_G_T_AC_C | Hap_3 | NA | NA | NA | |
| A_C_C_G_G_C_C_T_G_A_C_A_C_AC_C | Hap_4 | NA | NA | NA | |
| A_C_C_G_G_A_T_G_G_A_C_A_C_AC_C | Hap_5 | NA | NA | NA | |
| GH_D02G0972 | A_C_A_C_T_T_C_C_G_G_G | Hap_0 | a | a | a |
| A_C_A_C_T_T_C_C_G_A_G | Hap_1 | b | b | b | |
| T_T_G_A_T_T_A_C_A_G_T | Hap_2 | NA | NA | NA | |
| GH_A10G2325 | A_T_G_A_G_C_T | Hap_0 | a | a | a |
| A_T_G_A_G_C_C | Hap_1 | b | b | b | |
| T_C_A_T_T_T_C | Hap_2 | ab | c | b | |
| GH_D10G2421 | C_T_T_A_G_T_A_G_C_A_T_C_A_T_A_G_A_T_C_G_C_A_A_C_A_A_TC_A_T_C_C_T_G_G_T_A_T_A_G_A_A_G_G_TG_C_A_T_C_TCACTCAATGTGCTAATATGTCACATAACAATCCATTAAAAG AAAACAAAAAA_T_T_T_G_C_T_G_T_T_A_T_A_T_T_G_T_A_G_GA_C_C_G_G_G | Hap_0 | a | a | a |
| C_G_T_A_T_G_C_T_T_G_C_A_G_C_G_A_T_G_A_A_C_A_G_G_G_G_T_G_C_G_A_C_A_A_A_G_G_T_C_G_A_G_G_T_T_G_T_T_T_T_T_C_GTAC_T_A_A_C_G_G_C_A_A_G_A_C_T_C_G_A_G_G_G_G | Hap_1 | b | b | b | |
| C_T_T_A_G_T_A_G_C_A_T_C_A_C_A_G_A_T_C_G_C_A_A_C_A_A_TC_A_T_C_C_T_G_G_T_A_T_A_G_A_A_G_G_TG_C_A_T_C_TCACTCAATGTGCTAATATGTCACATAACAATCCATTAAAAGAAAA CAAAAAA_T_A_T_G_C_T_G_T_T_A_T_G_T_T_G_T_A_G_GA_C_C_G_G_A | Hap_2 | ab | ab | ab | |
| T_G_T_A_G_T_A_G_C_A_T_A_A_C_A_G_A_T_C_G_C_A_A_C_A_A_TC_A_T_C_C_T_G_G_T_A_T_A_G_A_A_G_G_TG_C_A_T_C_TCACTCAATGTGCTAATATGTCACATAACAATCCATTAAAAGAAAAC AAAAAA_T_T_T_G_C_T_G_T_T_A_T_A_T_T_G_T_A_G_GA_C_C_G_G_G | Hap_3 | NA | NA | NA | |
| C_T_T_A_G_T_A_G_C_A_T_C_A_C_A_G_A_T_C_G_C_A_A_C_A_A_TC_A_T_C_C_T_G_G_T_A_T_A_G_A_A_G_G_TG_C_A_T_C_TCACTCAATGTGCTAATATGTCACATAACAATCCATTAAAAGAAAAC AAAAAA_C_T_T_G_C_T_G_T_T_A_T_A_T_T_G_T_A_G_GA_C_C_G_G_A | Hap_4 | NA | NA | NA | |
| C_T_T_A_G_T_A_G_C_A_T_C_A_C_A_G_A_T_C_G_C_A_A_C_A_A_TC_A_T_C_C_T_G_G_T_A_T_A_G_A_A_G_G_TG_C_A_T_C_TCACTCAATGTGCTAATATGTCACATAACAATCCATTAAAAGAAAA CAAAAAA_T_T_T_G_C_T_G_T_T_A_T_A_T_T_G_T_A_G_GA_C_C_G_G_A | Hap_5 | NA | NA | NA | |
| GH_A12G2819 | G_A_G_G_C_C_A_G_T_G_CAGTGGGGACGAAGTT | Hap_0 | a | a | a |
| G_A_G_G_C_C_A_T_T_G_CAGTGGGGACGAAGTT | Hap_1 | b | b | b | |
| A_C_G_C_T_G_C_T_C_T_C | Hap_2 | NA | NA | NA | |
| G_C_T_G_C_G_A_T_C_G_CAGTGGGGACGAAGTT | Hap_3 | NA | NA | NA | |
| GH_A01G2253 | A_C_G_G_A_C_A_A_A_G_T_A_G_T_A_C | Hap_0 | a | a | a |
| A_T_G_A_C_C_T_A_G_C_TAA_A_A_C_A_C | Hap_1 | a | a | b | |
| C_C_G_A_A_G_T_A_G_C_T_T_A_C_A_A | Hap_2 | a | a | ab | |
| C_C_C_A_A_G_T_A_G_C_T_T_A_C_G_C | Hap_3 | a | a | ab | |
| A_C_G_A_A_C_A_T_G_C_T_A_A_C_A_C | Hap_4 | a | a | NA | |
| A_C_G_A_A_C_A_A_G_C_T_A_A_T_A_C | Hap_5 | a | a | NA | |
| GH_D08G2107 | C_G_T_G_A_C_A_G_A_A_T_G_T_T_A_A | Hap_0 | a | a | a |
| G_T_C_C_T_C_G_T_G_C_T_G_C_G_A_A | Hap_1 | b | b | b | |
| G_T_C_C_A_G_A_G_A_C_C_C_C_G_G_G | Hap_2 | NA | NA | NA | |
| G_T_C_C_A_G_A_G_A_C_T_C_C_G_G_G | Hap_3 | c | ab | ab | |
| GH_A08G2091 | A_A_T_C_C_G_C_T_C_G_C_T_AG_G_T_G_G_T_G_G_A | Hap_0 | a | a | a |
| A_A_T_C_C_G_T_T_C_G_C_T_AG_G_T_G_G_T_G_G_A | Hap_1 | b | b | a | |
| A_T_T_T_C_A_C_A_C_G_C_A_A_A_A_C_T_A_A_G_A | Hap_2 | ab | a | a | |
| A_A_T_C_T_A_C_T_C_G_C_T_A_A_A_C_T_A_A_G_A | Hap_3 | NA | NA | a | |
| A_A_T_T_C_A_C_T_C_G_C_A_A_A_A_C_T_A_A_G_A | Hap_4 | NA | NA | a | |
| A_A_T_C_C_G_C_T_C_G_C_T_AG_G_T_G_G_T_G_A_A | Hap_5 | ab | a | a | |
| C_A_A_C_C_A_C_T_T_T_A_T_AG_A_A_G_G_T_A_G_G | Hap_6 | ab | ab | a | |
| GH_D09G2155 | C_G_T_G | Hap_0 | a | a | a |
| A_T_T_G | Hap_1 | a | b | b | |
| C_G_T_T | Hap_2 | b | c | ab | |
| C_G_C_G | Hap_3 | NA | NA | NA | |
| A_G_T_G | Hap_4 | NA | NA | NA | |
| GH_D04G0442 | A_G_G_G_C_A_T_T_T | Hap_0 | a | a | a |
| A_G_A_G_C_G_T_C_T | Hap_1 | b | b | b | |
| T_A_G_G_G_G_T_T_G | Hap_2 | NA | NA | NA | |
| A_G_G_A_C_G_G_T_T | Hap_3 | NA | NA | NA | |
| A_G_G_G_C_G_T_T_T | Hap_4 | ab | ab | ab | |
| GH_A05G0964 | T_A_G_G_A_C_T_C_C_A_A_A_A_A_A_G | Hap_0 | a | a | a |
| T_A_G_G_T_C_C_C_C_A_A_A_A_A_A_G | Hap_1 | b | b | b | |
| A_A_G_T_A_T_T_A_T_T_T_A_T_C_AT_T | Hap_2 | NA | NA | NA | |
| T_G_T_G_A_C_T_C_C_A_A_T_A_A_A_G | Hap_3 | NA | NA | NA | |
| GH_D05G0951 | G_T_A_G | Hap_0 | a | a | a |
| A_C_G_T | Hap_1 | a | a | b | |
| G_T_A_T | Hap_2 | NA | NA | NA | |
| GH_A11G0638 | A_T_G_C_T_G_C_T_G_C_A_C_C_A_C | Hap_0 | a | a | a |
| G_T_A_G_C_A_C_A_G_T_A_A_C_G_C | Hap_1 | a | a | b | |
| A_A_G_G_T_G_C_T_A_C_C_C_G_A_T | Hap_2 | a | ab | ab | |
| G_T_A_G_C_G_C_A_G_T_A_A_C_G_C | Hap_3 | b | b | a | |
| A_A_G_G_T_G_T_T_A_C_C_C_G_A_T | Hap_4 | NA | NA | NA | |
| A_T_G_G_T_G_C_T_G_C_A_C_C_A_C | Hap_5 | a | ab | ab | |
| G_T_G_G_C_G_C_A_G_C_A_A_C_G_C | Hap_6 | b | ab | ab | |
| A_T_G_G_T_G_C_T_A_C_C_C_C_A_T | Hap_7 | NA | NA | NA | |
| GH_A08G1551 | T_A_C_G_G_T_C_T_A_T_C_T_G_T | Hap_0 | a | a | a |
| G_AT_C_A_A_G_A_C_G_G_T_C_C_G | Hap_1 | NA | NA | a | |
| T_A_A_G_G_T_C_T_A_T_C_T_G_T | Hap_2 | b | b | a | |
| GH_D08G1569 | T_A_T_G_C | Hap_0 | a | a | a |
| T_G_C_G_T | Hap_1 | b | a | a | |
| T_A_C_G_T | Hap_2 | NA | NA | NA | |
| A_A_T_T_C | Hap_3 | NA | NA | NA | |
| GH_D07G1883 | G_TAGA_G_C_T_G_T_C_T | Hap_0 | a | a | a |
| A_TAGA_G_T_A_A_C_A_T | Hap_1 | NA | NA | NA | |
| A_T_A_T_A_A_C_A_T | Hap_2 | a | b | b | |
| G_TAGA_G_C_T_G_T_C_C | Hap_3 | NA | NA | NA | |
| G_TAGA_G_C_T_A_T_C_T | Hap_4 | NA | NA | NA | |
| GH_A12G2452 | T_A_C_T_G_C_C_T_T_T_CT_T_C_G_TAAATTTTCTTAATTTCCGTTATTCAACCACAATATTTTTTAA_T | Hap_0 | a | a | a |
| C_G_C_C_G_C_C_C_T_T_C_T_C_A_TAAATTTTCTTAATTTCCGTTATTCAACCACAATATTTTTTAA_T | Hap_1 | a | b | b | |
| T_A_A_T_G_T_T_T_T_A_C_A_T_A_T_TTCAAAACGTTTGATGTAACCACTAA | Hap_2 | NA | NA | NA | |
| T_A_C_T_A_C_C_T_T_T_C_T_C_A_TAAATTTTCTTAATTTCCGTTATTCAACCACAATATTTTTTAA_T | Hap_3 | b | c | ab | |
| T_A_C_T_A_C_C_T_C_T_C_T_C_A_TAAATTTTCTTAATTTCCGTTATTCAACCACAATATTTTTTAA_T | Hap_4 | NA | NA | NA | |
| GH_A02G2044 | C_G_A_A_A_G_G_C_G_G_C_A_G_C_G_G_G | Hap_0 | a | a | a |
| C_G_A_G_A_G_G_C_G_G_C_A_G_T_G_G_G | Hap_1 | b | b | b | |
| T_G_G_G_G_C_G_T_T_C_T_G_A_C_G_G_A | Hap_2 | ab | ab | ab | |
| C_G_G_G_A_G_C_C_G_C_C_G_G_C_C_A_A | Hap_3 | ab | b | ab | |
| C_A_A_G_A_G_G_C_G_G_C_A_G_C_G_G_G | Hap_4 | NA | NA | NA |
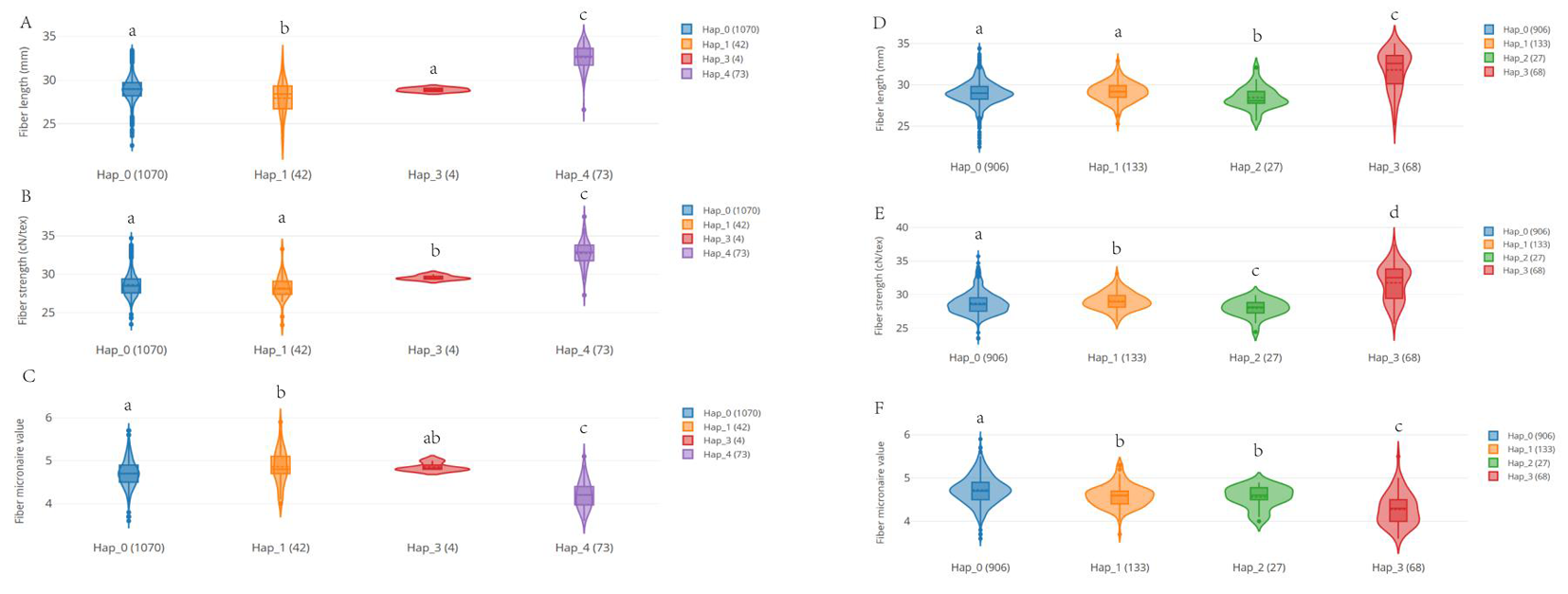
图8 部分陆地棉TRM基因单倍型与纤维品质关系 注:A:GH_A09G0827在群体中不同单倍型的材料纤维长度的比较;B:GH_A09G0827在群体中不同单倍型的材料纤维强度度的比较C:GH_A09G0827在群体中不同单倍型的材料马克隆值的比较;D:GH_D03G1434在群体中不同单倍型的材料纤维长度的比较;E:GH_D03G1434在群体中不同单倍型的材料纤维强度度的比较F:GH_D03G1434在群体中不同单倍型的材料马克隆值的比较。不同颜色代表着不同得单倍型。自上而下得三条线分别表示中位数和四分位距;Hap:不同的单倍型;多重比较字母标记法表示显著差异
Fig.8 Part of the relationship between Gossypium hirsutum L. TRM haplotype and fiber quality Note:A:Comparison of fiber length of GH_A09G0827 in different haplomorphic materials in population; B:Comparison of fiber strength of GH_A09G0827 materials with different haplotypes in the population C:comparison of Macron values of GH_A09G0827 materials with different haplotypes in the population; D:Comparison of fiber length of GH_D03G1434 in different haplomorphic materials in population; E:Comparison of fiber strength of GH_D03G1434 materials with different haplotypes in a population F:Comparison of Macron values of GH_D03G1434 materials with different haplotypes in a population.Different colors represent different haplotypes.Three lines from top to bottom represent the median and quartile distance respectively.Hap:Different haplotypes; Multiple comparison letter notation indicates significant differences
| [1] | 赵坤宇, 陈甜甜. 棉花转录因子SRS基因家族的鉴定与分析[J/OL]. 分子植物育种, 2022:1-11.(2022-08-04). https://kns.cnki.net/kcms/detail/46.1068.S.20220803.1704.014.html. |
| ZHAO Kunyu, CHEN Tiantian. Genome-wide identification and analysis of SRS gene family in cotton[J/OL]. Molecular Plant Breeding: 2022:1-11.(2022-08-04). https://kns.cnki.net/kcms/detail/46.1068.S.20220803.1704.014.html. | |
| [2] | 王坤波, 刘旭. 棉属多倍化研究进展[J]. 中国农业科技导报, 2013, 15(2):20-27. |
| WANG Kunbo, LIU Xu. Study progress of polyploidization[J]. Journal of Agricultural Science and Technology, 2013, 15(2):20-27. | |
| [3] |
Hu Y, Chen J D, Fang L, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton[J]. Nature Genetics, 2019, 51(4):739-748.
DOI |
| [4] |
Wang M J, Tu L L, Yuan D J, et al. Reference genome sequences of two cultivated allotetraploid cottons,Gossypium hirsutum L. and Gossypium barbadense[J]. Nature Genetics, 2019, 51(2):224-229.
DOI |
| [5] |
李雨哲, 邢淋雪, 刘梦洁, 等. 棉花苯丙氨酸解氨酶基因家族的生物信息学分析[J]. 棉花学报, 2021, 33(1):66-74.
DOI |
| LI Yuzhe, XING Linxue, LIU Mengjie, et al. Bioinformatics analysis of the phenylalanine ammonia lyase(PAL) gene family in cotton[J]. Cotton Science, 2021, 33(1):66-74. | |
| [6] | 李豪, 虎亚静, 张清霞, 等. 黄瓜TRM基因家族鉴定及分析[J]. 山东农业大学学报(自然科学版), 2021, 52(3):358-363. |
| LI Hao, HU Yajing, ZHANG Qingxia, et al. Identification and analysis on TRM family in cucumber[J]. Journal of Shandong Agricultural University(Natural Science Edition), 2021, 52(3):358-363. | |
| [7] |
Drevensek S, Goussot M, Duroc Y, et al. The Arabidopsis TRM1-TON1 interaction reveals a recruitment network common to plant cortical microtubule arrays and eukaryotic centrosomes[J]. The Plant Cell, 2012, 24(1):178-191.
DOI URL |
| [8] |
Guo Q Q, Ng P Q, Shi S S, et al. Arabidopsis TRM5 encodes a nuclear-localised bifunctional tRNA guanine and inosine-N1-methyltransferase that is important for growth[J]. PLoS One, 2019, 14(11):e0225064.
DOI URL |
| [9] |
Yang B, Voiniciuc C, Fu L B, et al. TRM4 is essential for cellulose deposition in Arabidopsis seed mucilage by maintaining cortical microtubule organization and interacting with CESA3[J]. The New Phytologist, 2019, 221(2):881-895.
DOI URL |
| [10] |
Schaefer E, Belcram K, Uyttewaal M, et al. The preprophase band of microtubules controls the robustness of division orientation in plants[J]. Science, 2017, 356(6334):186-189.
DOI PMID |
| [11] | Tang J, Jia P F, Xin P Y, et al. The Arabidopsis TRM61/TRM6 complex is a bona fide tRNA N1-methyladenosine methyltransferase[J]. Journal of Experi-mental Botany, 2020, 71(10):3024-3036. |
| [12] |
Qanmber G, YU D Q, LI J, et al. Genome-wide identification and expression analysis of Gossypium RING-H2 finger E3 ligase genes revealed their roles in fiber development,and phytohormone and abiotic stress responses[J]. Journal of Cotton Research, 2018, 1(1):1.
DOI |
| [13] |
YANG X, XU Y C, YANG F F, et al. Genome-wide identification of OSCA gene family and their potential function in the regulation of dehydration and salt stress in Gossypium hirsutum[J]. Journal of Cotton Research, 2019, 2(1):11.
DOI |
| [14] | 柳思思. 玉米耐旱功能标记开发及优异单倍型应用[D]. 乌鲁木齐: 新疆农业大学, 2012. |
| LIU Sisi. The Development of Functional Market and the Application of Superior Haplotype Associated with Drought Tolerance in Maize(zea Mays L.)[D]. Urumqi: Xinjiang Agricultural University, 2012. | |
| [15] |
Zhu T, Liang C, Meng Z G, et al. CottonFGD:an integrated functional genomics database for cotton[J]. BMC Plant Biology, 2017, 17(1):101.
DOI URL |
| [16] | Mount D W. Using the basic local alignment search tool(BLAST)[J]. Cold Spring Harbor Protocols, 2007, 2007(7):pdb.top17. |
| [17] |
Finn R D, Clements J, Eddy S R. HMMER web server:interactive sequence similarity searching[J]. Nucleic Acids Research, 2011, 39:W29-W37
DOI URL |
| [18] |
Chen C J, Chen H, Zhang Y, et al. TBtools:an integrative toolkit developed for interactive analyses of big biological data[J]. Molecular Plant, 2020, 13(8):1194-1202.
DOI URL |
| [19] |
Hall B G. Building phylogenetic trees from molecular data with MEGA[J]. Molecular Biology and Evolution, 2013, 30(5):1229-1235.
DOI PMID |
| [20] |
Larkin M A, Blackshields G, Brown N P, et al. Clustal W and Clustal X version 2.0.[J]. Bioinformatics, 2007, 23(21):2947-2948.
DOI PMID |
| [21] |
Lescot M. PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Research, 2002, 30(1):325-327.
DOI URL |
| [22] |
Ma Z Y, He S P, Wang X F, et al. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield[J]. Nature Genetics, 2018, 50(6):803-813.
DOI PMID |
| [23] |
He S P, Sun G F, Geng X L, et al. The genomic basis of geographic differentiation and fiber improvement in cultivated cotton[J]. Nature Genetics, 2021, 53(6):916-924.
DOI PMID |
| [24] |
Li F, Fan G, Lu C, et al. Genome sequence of cultivated Upland cotton(Gossypium hirsutum L. TM-1) provides insights into genome evolution[J]. Nat Biotechnol, 2015, 33(5):524-530.
DOI |
| [25] |
Zhang T Z, Hu Y, Jiang W K, et al. Sequencing of allotetraploid cotton(Gossypium hirsutum L.acc.TM-1) provides a resource for fiber improvement[J]. Nature Biotechnology, 2015, 33(5):531-537.
DOI |
| [26] |
Li F, Fan G, Wang K, et al. Genome sequence of the cultivated cotton Gossypium arboreum[J]. Nat Genet, 2014, 46(6):567-72.
DOI PMID |
| [27] |
Ma J J, Jiang Y F, Pei W F, et al. Expressed genes and their new alleles identification during fibre elongation reveal the genetic factors underlying improvements of fibre length in cotton[J]. Plant Biotechnology Journal, 2022, 20(10):1940-1955.
DOI URL |
| [1] | 张庭军, 李字辉, 崔豫疆, 孙孝贵, 陈芳. 微生物菌剂对棉花生长及土壤理化性质的影响[J]. 新疆农业科学, 2024, 61(9): 2269-2276. |
| [2] | 李颖, 郭文文, 李江博, 曲延英, 陈全家, 郑凯. 90份转BT基因抗虫棉品种(系)在新疆早熟棉区的适应性评价[J]. 新疆农业科学, 2024, 61(7): 1561-1573. |
| [3] | 杨彩霞, 顾炜, 关媛, 瞿静涛, 党冬冬, 吴鹏昊, 郑洪建. 甜玉米基因Sugary1(Su1)序列的变异分析[J]. 新疆农业科学, 2024, 61(7): 1605-1614. |
| [4] | 巩隽铭, 熊显鹏, 张彩霞, 邵东南, 程帅帅, 孙杰. 陆地棉4-香豆酸辅酶A连接酶基因Gh4CL30的功能分析[J]. 新疆农业科学, 2024, 61(6): 1301-1309. |
| [5] | 马尚洁, 李生梅, 杨涛, 王红刚, 赵康, 庞博, 高文伟. 陆地棉GHWAT1-35基因的克隆及亚细胞定位[J]. 新疆农业科学, 2024, 61(6): 1310-1317. |
| [6] | 董祯林, 万素梅, 熊世武, 马云珍, 毛廷勇, 杨北方, 骆磊, 刘超群, 陈国栋, 李亚兵. 不同种植密度对中棉113农艺性状及产量的影响[J]. 新疆农业科学, 2024, 61(5): 1102-1111. |
| [7] | 刘太杰, 陈兵, 杨立, 王静, 赵静, 李翔, 唐广兰, 王刚, 韩焕勇, 王方永. 不同喷药机械与药剂组合对棉花脱叶催熟效果及产量和品质的影响[J]. 新疆农业科学, 2024, 61(4): 852-860. |
| [8] | 张伟, 杨国慧, 于辉. 2,4-表油菜素内酯对干旱胁迫下西瓜幼苗生长及相关基因表达的影响[J]. 新疆农业科学, 2024, 61(3): 615-622. |
| [9] | 崔豫疆, 龚照龙, 王俊铎, 郑巨云, 桑志伟, 阳妮, 梁亚军, 李雪源, 曲延英. 245份陆地棉品种农艺性状及产量构成因素综合评价[J]. 新疆农业科学, 2024, 61(10): 2358-2365. |
| [10] | 赵康, 任丹, 梁维维, 庞博, 马尚洁, 张梦媛, 高文伟. 陆地棉正反交F2∶3家系主要农艺性状与单株皮棉产量的关系[J]. 新疆农业科学, 2024, 61(1): 19-25. |
| [11] | 王朋, 郑凯, 赵杰银, 高文举, 龙遗磊, 陈全家, 曲延英. 陆地棉种质资源材料的耐热性评价及指标筛选[J]. 新疆农业科学, 2023, 60(9): 2081-2090. |
| [12] | 王辉, 郭金成, 宋佳, 张庭军, 何良荣. 高温胁迫下陆地棉GhCIPK6转基因后代生理生化分析[J]. 新疆农业科学, 2023, 60(9): 2109-2119. |
| [13] | 马青山, 杜霄, 陶志鑫, 韩万里, 龙遗磊, 艾先涛, 胡守林. 陆地棉种质材料机采农艺性状鉴定分析[J]. 新疆农业科学, 2023, 60(8): 1830-1839. |
| [14] | 段松江, 彭增莹, 申莹莹, 木丽迪尔·拜波拉提, 吴一凡, 崔建平, 张巨松. 不同海岛棉品种产量及纤维品质对氮肥的响应[J]. 新疆农业科学, 2023, 60(7): 1569-1579. |
| [15] | 邵盘霞, 赵准, 邵武奎, 郝晓燕, 高升旗, 李建平, 胡文冉, 黄全生. 玉米ZmCDPK22基因在干旱胁迫下的表达分析[J]. 新疆农业科学, 2023, 60(6): 1372-1378. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||