

新疆农业科学 ›› 2024, Vol. 61 ›› Issue (6): 1301-1309.DOI: 10.6048/j.issn.1001-4330.2024.06.001
• 作物遗传育种•种质资源•分子遗传学•耕作栽培•生理生化 • 上一篇 下一篇
巩隽铭( ), 熊显鹏, 张彩霞, 邵东南, 程帅帅, 孙杰(
), 熊显鹏, 张彩霞, 邵东南, 程帅帅, 孙杰( )
)
收稿日期:2023-10-23
出版日期:2024-06-20
发布日期:2024-08-08
通信作者:
孙杰(1969-),男,新疆人,教授,博士,硕士生/博士生导师,研究方向为棉花遗传育种,(E-mail) sunjie@shzu.edu.cn作者简介:巩隽铭(1995- ),男,辽宁人,硕士研究生,研究方向为棉花分子育种,(E-mail) gongjunming_cn@163.com
基金资助:
GONG Junming( ), XIONG Xianpeng, ZHANG Caixia, SHAO Dongnan, CHENG Shuaishuai, SUN Jie(
), XIONG Xianpeng, ZHANG Caixia, SHAO Dongnan, CHENG Shuaishuai, SUN Jie( )
)
Received:2023-10-23
Published:2024-06-20
Online:2024-08-08
Correspondence author:
SUN Jie (1969-), male, from Xinjiang, professor,Ph.D, research direction: cotton genetics and breeding, (E-mail) sunjie@shzu.edu.cnSupported by:摘要:
【目的】研究4-香豆酸辅酶A连接酶(4CL)家族基因Gh4CL30的生物学功能,为棉花株型育种提供理论依据和基因种质资源。【方法】利用病毒诱导的基因沉默技术和基因编辑技术获得Gh4CL30沉默和编辑植株,测定该基因抑制及敲除植株的黄酮和木质素含量,调查棉花田间表型性状、种子大小和纤维品质。【结果】Gh4CL30基因沉默和敲除植株中木质素合成相关基因表达量显著下降,4-香豆酸辅酶A连接酶含量显著减少,茎秆中木质素含量显著降低,其株高、种子大小和纤维长度显著降低。【结论】Gh4CL30通过调控木质素生物合成功能影响棉花的生长发育。
中图分类号:
巩隽铭, 熊显鹏, 张彩霞, 邵东南, 程帅帅, 孙杰. 陆地棉4-香豆酸辅酶A连接酶基因Gh4CL30的功能分析[J]. 新疆农业科学, 2024, 61(6): 1301-1309.
GONG Junming, XIONG Xianpeng, ZHANG Caixia, SHAO Dongnan, CHENG Shuaishuai, SUN Jie. Functional analysis of 4-coumarate: CoA ligase gene Gh4CL30 in upland cotton[J]. Xinjiang Agricultural Sciences, 2024, 61(6): 1301-1309.
| 基因名称 Genes Name | 基因ID Gene ID | 染色体 Chromosome | 蛋白长度 Protein Length (aa) | 等电点 Isoelectric Point | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| Gh4CL6 | GH_A05G0051 | A05 | 543 | 5.835 | Plasma membrane |
| Gh4CL7 | GH_A05G1439 | A05 | 543 | 6.09 | cytosol |
| Gh4CL12 | GH_A10G0499 | A09 | 129 | 10.388 | chloroplast |
| Gh4CL24 | GH_D05G0056 | D05 | 540 | 6.25 | chloroplast |
| Gh4CL25 | GH_D05G1456 | D05 | 543 | 5.872 | cytosol |
| Gh4CL30 | GH_D10G0525 | D10 | 543 | 5.284 | cytosol |
表1 陆地棉中的Ⅰ型Gh4CL基因
Tab.1 Type I Gh4CL genes in G.hirsutum
| 基因名称 Genes Name | 基因ID Gene ID | 染色体 Chromosome | 蛋白长度 Protein Length (aa) | 等电点 Isoelectric Point | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| Gh4CL6 | GH_A05G0051 | A05 | 543 | 5.835 | Plasma membrane |
| Gh4CL7 | GH_A05G1439 | A05 | 543 | 6.09 | cytosol |
| Gh4CL12 | GH_A10G0499 | A09 | 129 | 10.388 | chloroplast |
| Gh4CL24 | GH_D05G0056 | D05 | 540 | 6.25 | chloroplast |
| Gh4CL25 | GH_D05G1456 | D05 | 543 | 5.872 | cytosol |
| Gh4CL30 | GH_D10G0525 | D10 | 543 | 5.284 | cytosol |
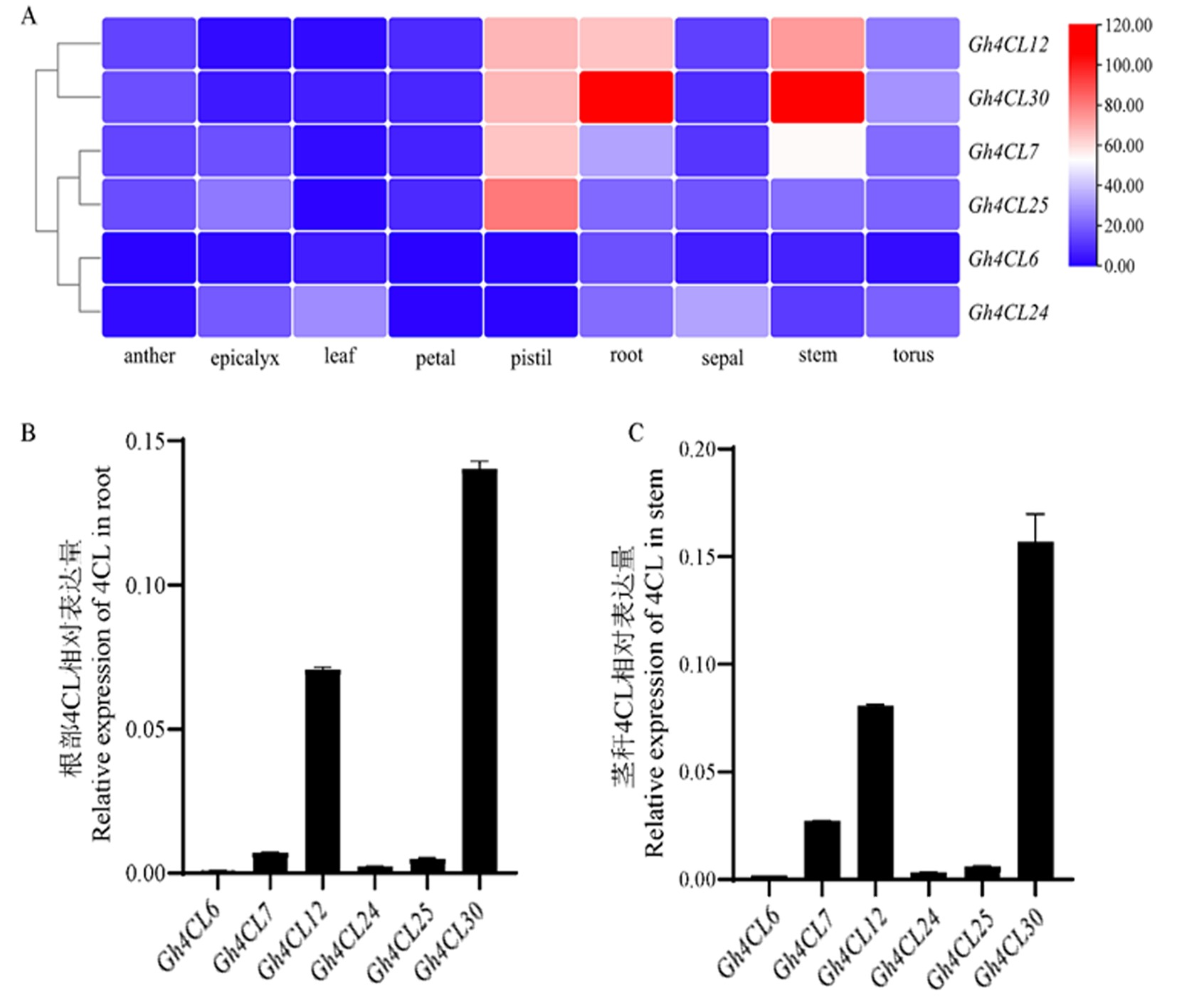
图1 6个Ⅰ型Gh4CL基因在棉花不同组织中的表达模式 注:A:6个Ⅰ型Gh4CL基因在棉花不同组织中的表达模式;B:6个Ⅰ型Gh4CL基因在棉花根中的相对表达量;C:6个Ⅰ型Gh4CL基因在棉花茎杆中的相对表达量
Fig.1 Expression patterns of six type I Gh4CL in different tissues of cotton Note: A:Expression patterns of six type I Gh4CLs in different tissues of cotton;B:Relative expression of six type I Gh4CL genes in cotton roots;C:Relative expression of six type I Gh4CL genes in cotton stems
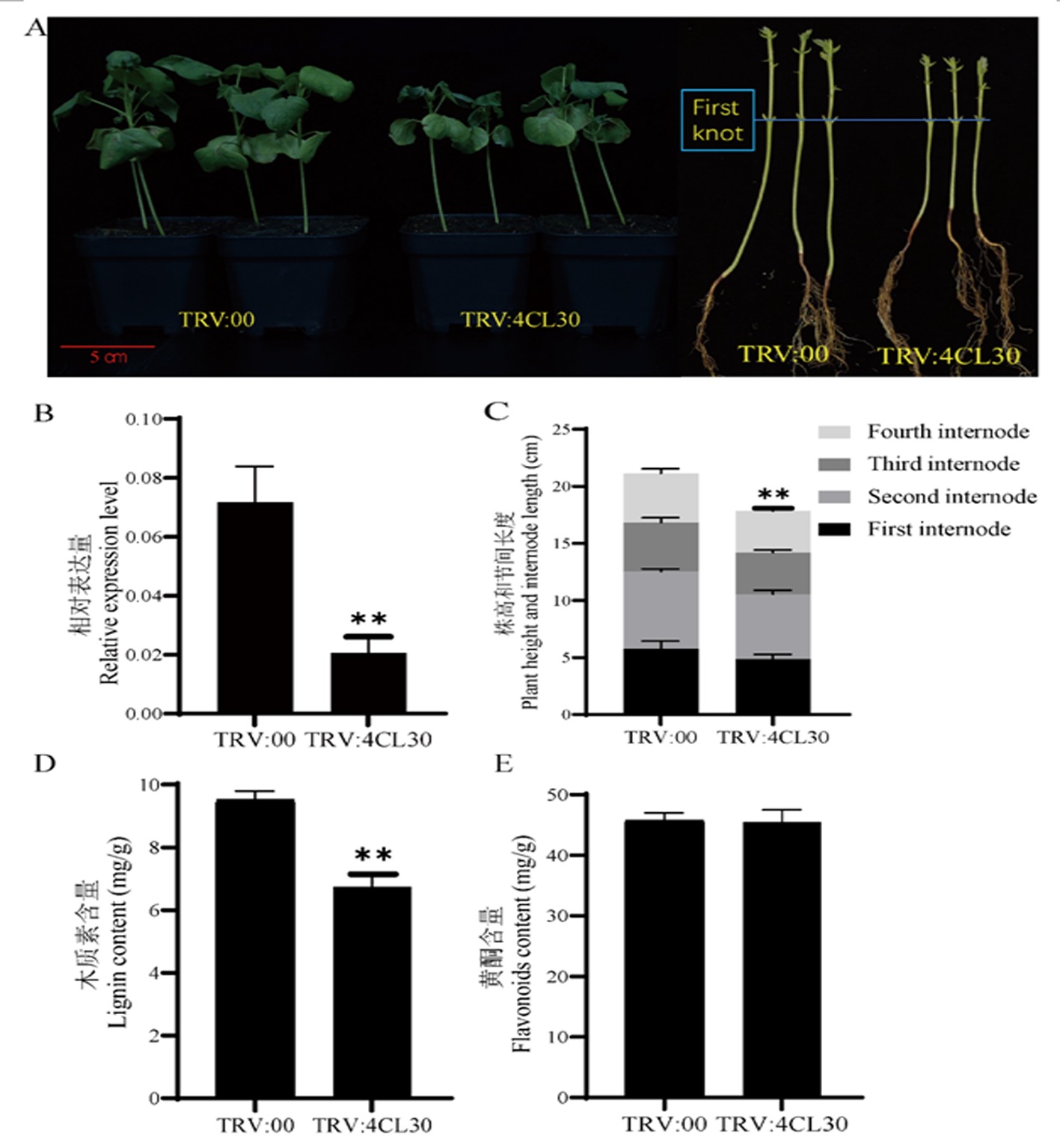
图2 Gh4CL30基因沉默植株的表型和相关指标测定 注:A:TRV:00和TRV:Gh4CL30植株之间的表型差异;B:Gh4CL30基因在TRV:00和TRV:Gh4CL30植株中的相对表达量;C:TRV:00和TRV:Gh4CL30植株之间的株高和节间长度差异,由下到上依次是棉花植株的第1至4节间长度;D:TRV:00和TRV:Gh4CL30植株的木质素含量差异;E:TRV:00和TRV:Gh4CL30植株的黄酮类化合物含量差异
Fig.2 Phenotype and determination of relevant indicators in Gh4CL30 gene silenced plants Note: A:Phenotypic differences between TRV:Gh4CL30 and TRV:00 plants;B:Relative expression of Gh4CL30 in TRV:00 and TRV:Gh4CL30 plants;C:Differences in plant height and internode length between TRV:00 and TRV:Gh4CL30 plants.From bottom to top, the first to fourth internode lengths of cotton plants are shown;D:Lignin content of TRV:00 and TRV:Gh4CL30 plants;E:Flavonoid metabolite content of TRV:00 and TRV:Gh4CL30 plants
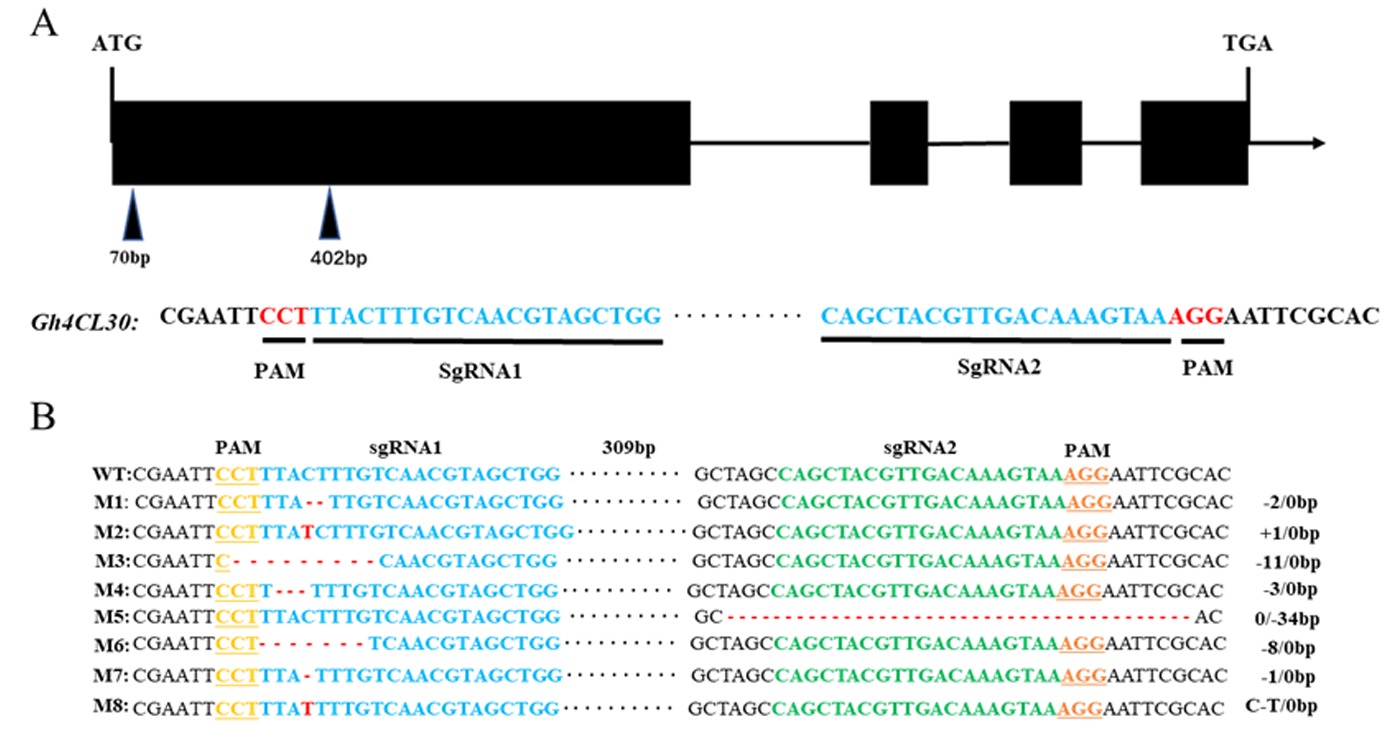
图3 Gh4CL30基因结构和编辑靶标的选择以及基因编辑类型 注:A:在Gh4CL30基因的第一个外显子区域中选择了两个编辑靶标;B:突变体的基因编辑类型
Fig.3 Gh4CL30 gene structure and selection of editing targets and types of gene editing Note:A:Two editing targets were selected in the first exon region of the Gh4CL30 gene;B:Type of gene editing of the mutant
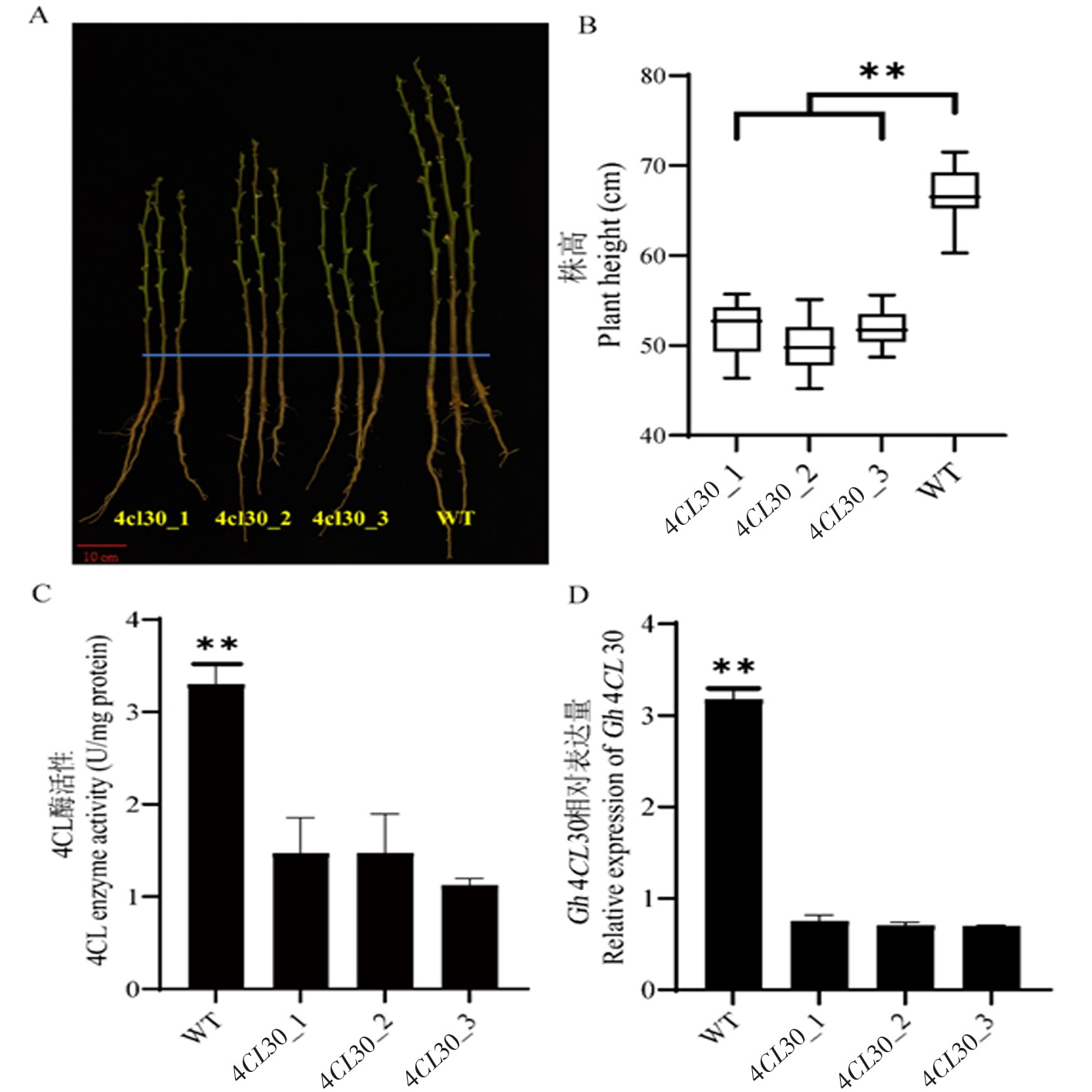
图4 WT和4CL30植株的表型、4CL酶活性以及Gh4CL30的相对表达量 注:A:4CL30编辑植株和对照WT株高的表型比较;B:4CL30编辑植株和对照WT株高统计;C:对照WT和4CL30植株的4CL酶活性差异;D:WT和4CL30编辑植株茎秆中Gh4CL30基因相表达量
Fig.4 Phenotype, 4CL enzyme activity and relative expression of Gh4CL30 in WT and 4CL30 plants Note: A:Phenotypic comparison of 4CL30 and WT plant heights;B:Plant height statistics of 4CL30 edited plants and WT; C:The 4CL enzyme activity of WT and 4CL30 plants was different;D:Relative expression of Gh4CL30 gene in stems of WT and 4CL30 edited plants
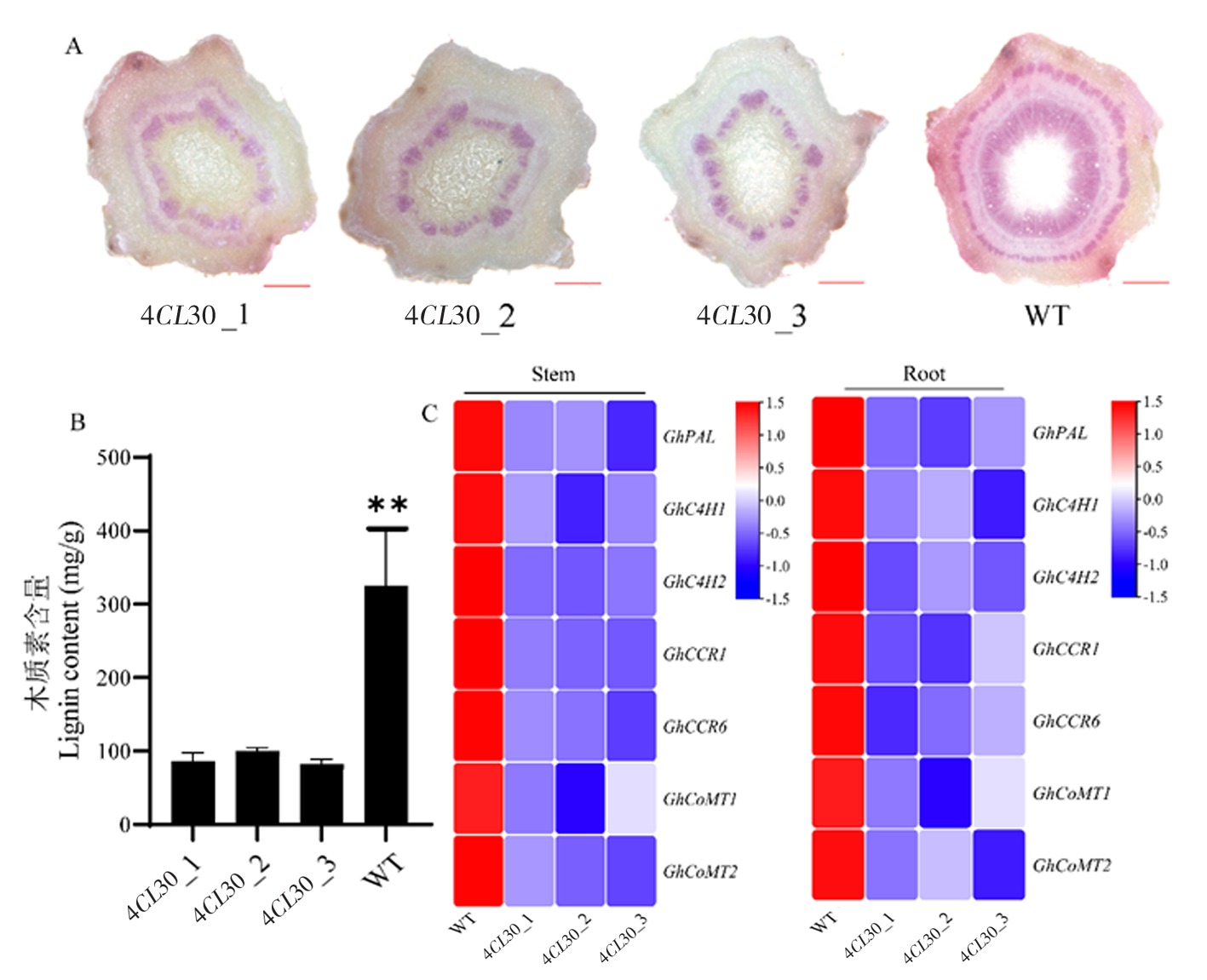
图5 4CL30和WT植株的木质素含量以及木质素合成相关基因的表达量 注:A:使用盐酸和间苯三酚对4CL30和对照WT植物在同一节点的茎段切片染色;B:4CL30和对照WT植株的木质素含量测定;C:4CL30和对照WT植株茎秆和根部组织中木质素合成相关基因的表达量
Fig.5 Lignin content of 4CL30 and WT plants and expression of genes related to lignin synthesis Note:A:Staining of stem sections of 4CL30 and WT plants at the same node using hydrochloric acid-phloroglucinol;B:Determination of lignin content in 4CL30 and WT plants;C:Expression of genes related to lignin synthesis in stem and root tissues of 4CL30 and WT plants
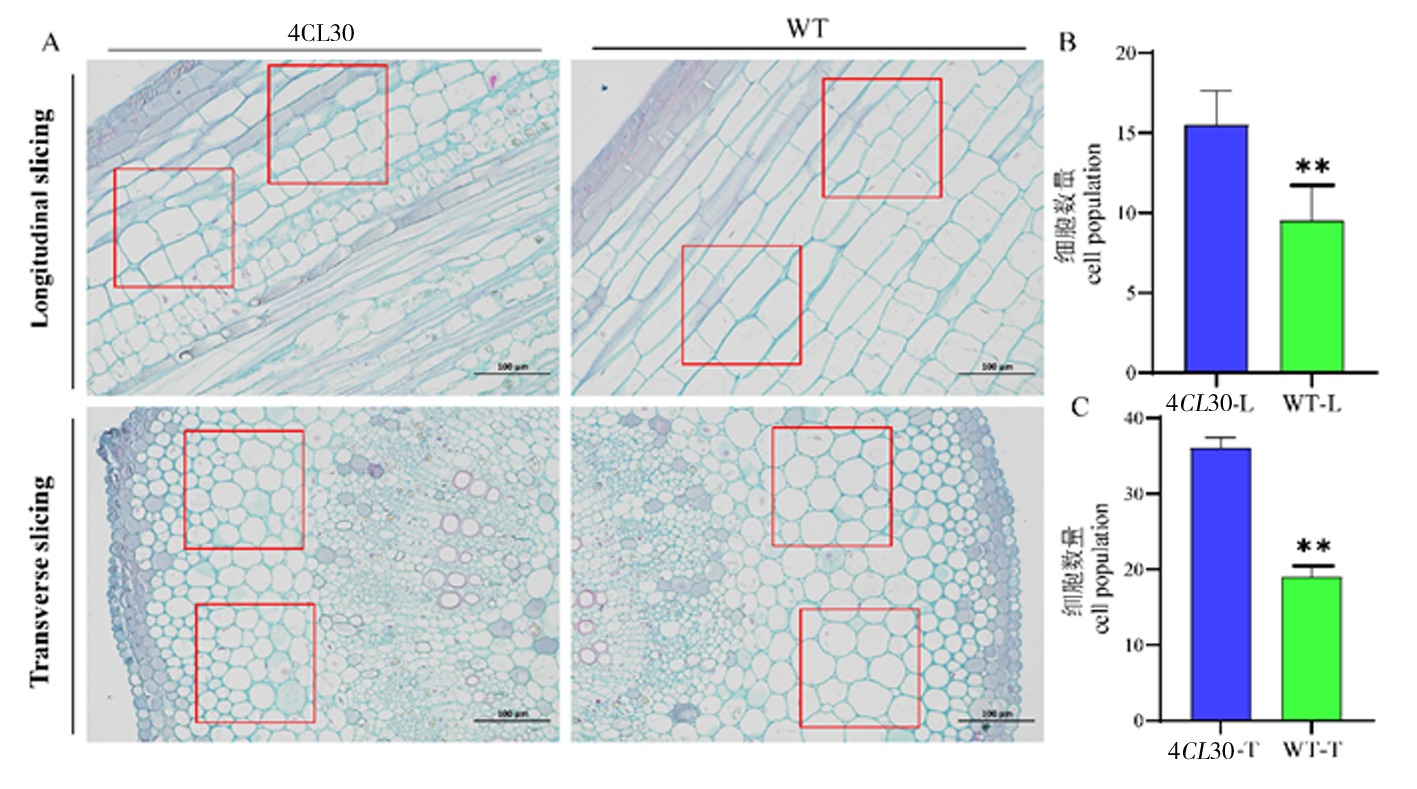
图6 4CL30和对照WT植株的细胞学观察 注:A:4CL30和对照WT植物相同节茎的横向和纵向石蜡切片;B:纵向切片中的细胞计数;C:横向切片中的细胞计数(同一比例下,对相同大小的红框中的细胞进行计数;细胞数量少则表示细胞长度较长,细胞大小较大。)
Fig.6 Cytological observations of 4CL30 and WT plants Note:A:Transverse and longitudinal paraffin sections of same-node stems of 4CL30 and WT plants;B:Cell counts in longitudinal sections;C:Cell counting in transverse sections (Cells in red boxes of the same size are counted at the same scale; a small number of cells indicates a longer cell length and a larger cell size.)
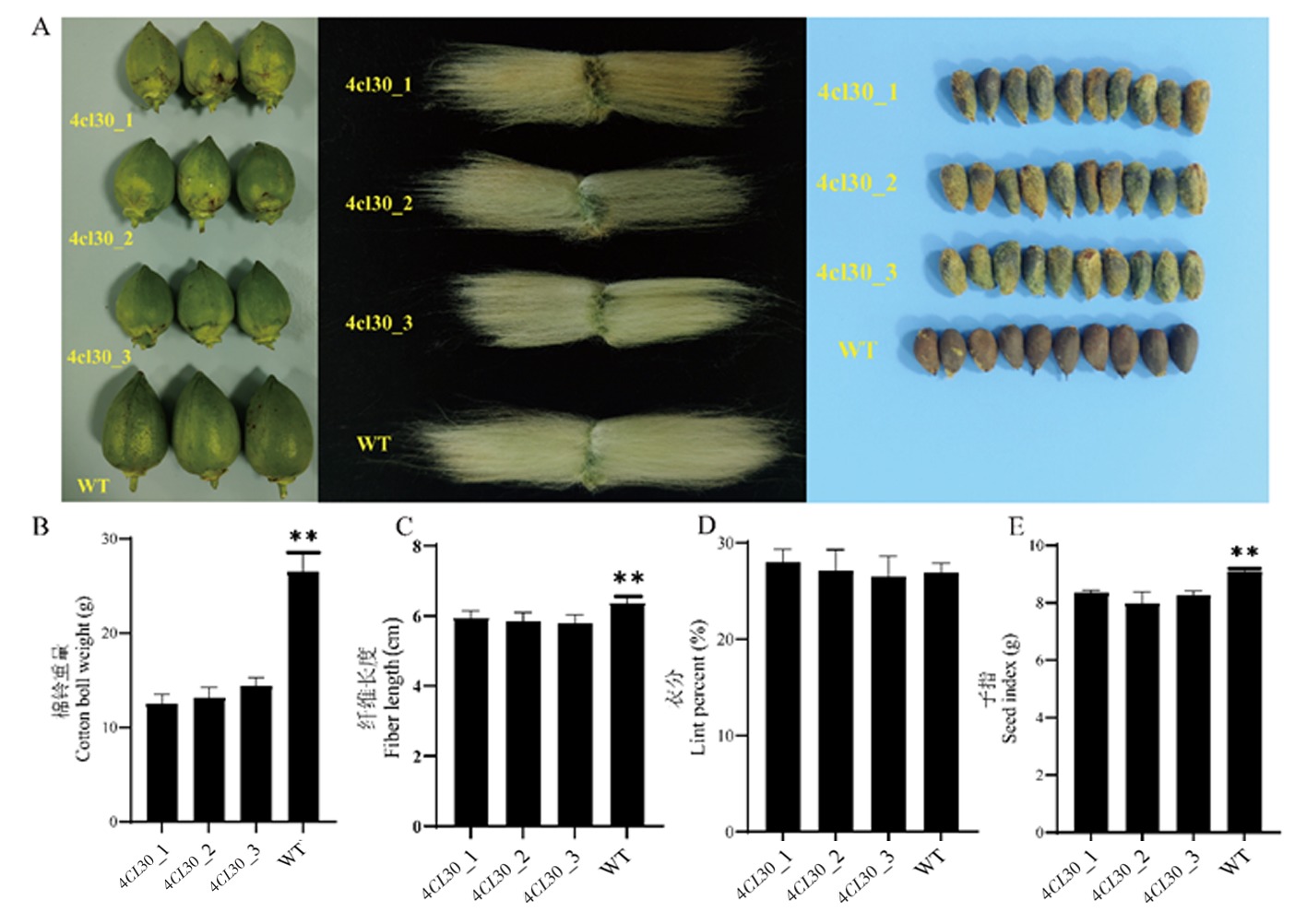
图7 对照WT和4CL30棉铃大小和纤维长度等表型比较 注:A:从左到右依次是植株棉铃大小、纤维长度和种子大小;B:4CL30和对照WT植株的棉铃重量(同一时期第一果枝上的棉铃);C:4CL30和对照WT植株的纤维长度;D:4CL30和对照WT的衣分;E: 4CL30和对照WT的子指
Fig.7 Phenotypic comparison of boll size and fiber length of WT and 4CL30 plants Note:A:From left to right are plant boll size, fiber length and seed size;B:Cotton boll weight (Cotton bolls on the first fruit branch of the same period) of WT and 4CL30 plants;C:Fiber length of WT and 4CL30 plants;D:Lint percent of WT and 4CL30 plants;E:Seed index of WT and 4CL30 plants
| [1] | Sun Q, Xie Y H, Li H M, et al. Cotton GhBRC1 regulates branching, flowering, and growth by integrating multiple hormone pathways[J]. The Crop Journal, 2022, 10(1): 75-87. |
| [2] | Bensen R J, Johal G S, Crane V C, et al. Cloning and characterization of the maize An1 gene[J]. The Plant Cell, 1995, 7(1): 75-84. |
| [3] |
Fu J Y, Ren F, Lu X, et al. A tandem array of ent-kaurene synthases in maize with roles in gibberellin and more specialized metabolism[J]. Plant Physiology, 2016, 170(2): 742-751.
DOI PMID |
| [4] |
Vogt T. Phenylpropanoid biosynthesis[J]. Molecular Plant, 2010, 3(1): 2-20.
DOI PMID |
| [5] |
Wang S C, Alseekh S, Fernie A R, et al. The structure and function of major plant metabolite modifications[J]. Molecular Plant, 2019, 12(7): 899-919.
DOI PMID |
| [6] |
Zhao Q. Lignification: flexibility, biosynthesis and regulation[J]. Trends in Plant Science, 2016, 21(8): 713-721.
DOI PMID |
| [7] |
Nakabayashi R, Saito K. Integrated metabolomics for abiotic stress responses in plants[J]. Current Opinion in Plant Biology, 2015, 24: 10-16.
DOI PMID |
| [8] |
Le Roy J, Huss B, Creach A, et al. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants[J]. Frontiers in Plant Science, 2016, 7: 735.
DOI PMID |
| [9] |
Amrhein N, Frank G, Lemm G, et al. Inhibition of lignin formation by L-alpha-aminooxy-beta-phenylpropionic acid, an inhibitor of phenylalanine ammonia-lyase[J]. European Journal of Cell Biology, 1983, 29(2): 139-144.
PMID |
| [10] | Smart C C, Amrhein N. The influence of lignification on the development of vascular tissue inVigna radiata L[J]. Protoplasma, 1985, 124(1): 87-95. |
| [11] |
Lavhale S G, Kalunke R M, Giri A P. Structural, functional and evolutionary diversity of 4-coumarate-CoA ligase in plants[J]. Planta, 2018, 248(5): 1063-1078.
DOI PMID |
| [12] | Voelker S L, Lachenbruch B, Meinzer F C, et al. Reduced wood stiffness and strength, and altered stem form, in young antisense 4CL transgenic poplars with reduced lignin contents[J]. The New Phytologist, 2011, 189(4): 1096-1109. |
| [13] | Chen X H, Wang H T, Li X Y, et al. Molecular cloning and functional analysis of 4-Coumarate: CoA ligase 4(4CL-like 1)from Fraxinus mandshurica and its role in abiotic stress tolerance and cell wall synthesis[J]. BMC Plant Biology, 2019, 19(1): 231. |
| [14] | Li Y, Kim J I, Pysh L, et al. Four isoforms of Arabidopsis 4-coumarate: CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism[J]. Plant Physiology, 2015, 169(4): 2409-2421. |
| [15] | Gui J S, Shen J H, Li L G. Functional characterization of evolutionarily divergent 4-coumarate: coenzyme a ligases in rice[J]. Plant Physiology, 2011, 157(2): 574-586. |
| [16] | Shi R, Sun Y H, Li Q Z, et al. Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes[J]. Plant & Cell Physiology, 2010, 51(1): 144-163. |
| [17] | Ehlting J, Büttner D, Wang Q, et al. Three 4-coumarate: coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms[J]. The Plant Journal, 1999, 19(1): 9-20. |
| [18] |
Wang B, Sun W, Li Q S, et al. Genome-wide identification of phenolic acid biosynthetic genes in Salvia miltiorrhiza[J]. Planta, 2015, 241(3): 711-725.
DOI PMID |
| [19] | Sun S C, Xiong X P, Zhang X L, et al. Characterization of the Gh4CL gene family reveals a role of Gh4CL7 in drought tolerance[J]. BMC Plant Biology, 2020, 20(1): 125. |
| [20] | Zhang T Z, Hu Y, Jiang W K, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement[J]. Nature Biotechnology, 2015, 33(5): 531-537. |
| [21] |
Chen C J, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Molecular Plant, 2020, 13(8): 1194-1202.
DOI PMID |
| [22] | Gao W, Long L, Zhu L F, et al. Proteomic and virus-induced gene silencing (VIGS) Analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae[J]. Molecular & Cellular Proteomics, 2013, 12(12): 3690-3703. |
| [23] | Cheng X Q, Zhang X Y, Xue F, et al. Characterization and transcriptome analysis of a dominant genic male sterile cotton mutant[J]. BMC Plant Biology, 2020, 20(1): 312. |
| [24] | Fan L, Shi W J, Hu W R, et al. Molecular and biochemical evidence for phenylpropanoid synthesis and presence of wall-linked phenolics in cotton fibers[J]. Journal of Integrative Plant Biology, 2009, 51(7): 626-637. |
| [25] | 付远志, 薛惠云, 胡根海, 等. 我国棉花株型性状遗传育种研究进展[J]. 江苏农业科学, 2019, 47(5): 16-19. |
| FU Yuanzhi, XUE Huiyun, HU Genhai, et al. Research progress on genetics breeding of plant architecture traits of cotton in China[J]. Jiangsu Agricultural Sciences, 2019, 47(5): 16-19. | |
| [26] | Li X, Bonawitz N D, Weng J K, et al. The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids[J]. The Plant Cell, 2010, 22(5): 1620-1632. |
| [27] |
Hu W J, Kawaoka A, Tsai C J, et al. Compartmentalized expression of two structurally and functionally distinct 4-coumarate: CoA ligase genes in aspen (Populus tremuloides)[J]. Proceedings of the National Academy of Sciences of the United States of America, 1998, 95(9): 5407-5412.
DOI PMID |
| [28] | Sutela S, Hahl T, Tiimonen H, et al. Phenolic compounds and expression of 4CL genes in silver birch clones and Pt4CL1a lines[J]. PLoS One, 2014, 9(12): e114434. |
| [29] |
Allina S M, Pri-Hadash A, Theilmann D A, et al. 4-Coumarate: coenzyme A ligase in hybrid poplar. Properties of native enzymes, cDNA cloning, and analysis of recombinant enzymes[J]. Plant Physiology, 1998, 116(2): 743-754.
PMID |
| [30] | Zhong R Q, Cui D T, Ye Z H. Secondary cell wall biosynthesis[J]. The New Phytologist, 2019, 221(4): 1703-1723. |
| [31] | Gou J Y, Wang L J, Chen S P, et al. Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis[J]. Cell Research, 2007, 17(5): 422-434. |
| [1] | 李颖, 郭文文, 李江博, 曲延英, 陈全家, 郑凯. 90份转BT基因抗虫棉品种(系)在新疆早熟棉区的适应性评价[J]. 新疆农业科学, 2024, 61(7): 1561-1573. |
| [2] | 卡地尔阿依·买买提, 周婷婷, 韩盛, 梅丽克汗·热西提, 玉山江·麦麦提. 不同甜瓜品种遗传转化再生体系的建立与基因编辑植株的快速获取[J]. 新疆农业科学, 2024, 61(7): 1666-1672. |
| [3] | 马尚洁, 李生梅, 杨涛, 王红刚, 赵康, 庞博, 高文伟. 陆地棉GHWAT1-35基因的克隆及亚细胞定位[J]. 新疆农业科学, 2024, 61(6): 1310-1317. |
| [4] | 王凯迪, 高晨旭, 裴文锋, 杨书贤, 张文庆, 宋吉坤, 马建江, 王莉, 于霁雯, 陈全家. 陆地棉TRM基因家族的鉴定及纤维品质相关优异单倍型分析[J]. 新疆农业科学, 2024, 61(3): 521-536. |
| [5] | 岳成广, 李忠慧, 刘晨曦, 贺三刚, 马海叶, 刘璇, 李婧平, 李文蓉. 不同基因编辑类型对FGF5基因编辑羊羊毛性状的影响[J]. 新疆农业科学, 2024, 61(3): 734-741. |
| [6] | 崔豫疆, 龚照龙, 王俊铎, 郑巨云, 桑志伟, 阳妮, 梁亚军, 李雪源, 曲延英. 245份陆地棉品种农艺性状及产量构成因素综合评价[J]. 新疆农业科学, 2024, 61(10): 2358-2365. |
| [7] | 赵康, 任丹, 梁维维, 庞博, 马尚洁, 张梦媛, 高文伟. 陆地棉正反交F2∶3家系主要农艺性状与单株皮棉产量的关系[J]. 新疆农业科学, 2024, 61(1): 19-25. |
| [8] | 王朋, 郑凯, 赵杰银, 高文举, 龙遗磊, 陈全家, 曲延英. 陆地棉种质资源材料的耐热性评价及指标筛选[J]. 新疆农业科学, 2023, 60(9): 2081-2090. |
| [9] | 王辉, 郭金成, 宋佳, 张庭军, 何良荣. 高温胁迫下陆地棉GhCIPK6转基因后代生理生化分析[J]. 新疆农业科学, 2023, 60(9): 2109-2119. |
| [10] | 马青山, 杜霄, 陶志鑫, 韩万里, 龙遗磊, 艾先涛, 胡守林. 陆地棉种质材料机采农艺性状鉴定分析[J]. 新疆农业科学, 2023, 60(8): 1830-1839. |
| [11] | 耿翡翡, 孟超敏, 卿桂霞, 周佳敏, 张富厚, 刘逢举. 陆地棉磷高效基因GhMYB4的克隆与表达分析[J]. 新疆农业科学, 2023, 60(6): 1406-1412. |
| [12] | 李佩琪, 孙庆培, 王志慧, 秦新政, 樊永红. 棉秆固体发酵中木质素降解与酶活性变化的关联分析[J]. 新疆农业科学, 2023, 60(6): 1423-1432. |
| [13] | 陈亮亮, 张梦, 郭立平, 戚廷香, 张学贤, 唐会妮, 王海林, 乔秀琴, 吴建勇, 邢朝柱. 陆地棉杂交组合F1、F2苗期优势表现及亲本配合力分析[J]. 新疆农业科学, 2023, 60(2): 261-271. |
| [14] | 龙天宇, 邓亚辉, 祖倩丽, 杨龙, 曲延英, 陈全家. 棉花枯萎病室内苗期抗病性鉴定方法及评价[J]. 新疆农业科学, 2023, 60(2): 416-423. |
| [15] | 文佳, 黄陈珏, 嵇子涵, 李黎贝, 冯震, 喻树迅. 陆地棉动态株高与SSR标记的关联分析[J]. 新疆农业科学, 2023, 60(12): 2892-2901. |
| 阅读次数 | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
全文 73
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
摘要 243
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||